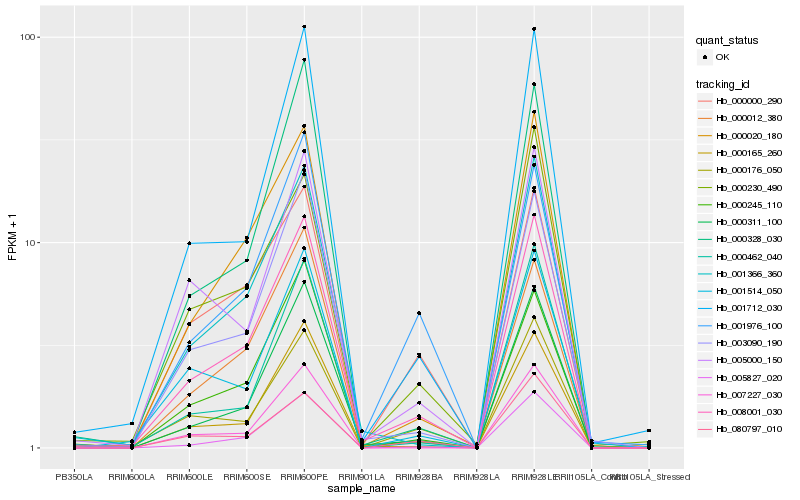
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008001_030 |
0.0 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000311_100 |
0.0754116368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000020_180 |
0.078815391 |
- |
- |
laccase, putative [Ricinus communis] |
| 4 |
Hb_001366_360 |
0.0808392019 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_001712_030 |
0.0858666451 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 19 [Jatropha curcas] |
| 6 |
Hb_005827_020 |
0.0859263216 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Populus euphratica] |
| 7 |
Hb_003090_190 |
0.0869543575 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 8 |
Hb_000165_260 |
0.0906186175 |
- |
- |
PREDICTED: uncharacterized protein LOC105133752 isoform X2 [Populus euphratica] |
| 9 |
Hb_000245_110 |
0.1005077158 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |
| 10 |
Hb_005000_150 |
0.1032280686 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001514_050 |
0.1059520318 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 12 |
Hb_007227_030 |
0.1069068896 |
- |
- |
hypothetical protein JCGZ_24975 [Jatropha curcas] |
| 13 |
Hb_000328_030 |
0.1089106374 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |
| 14 |
Hb_000012_380 |
0.1129172234 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_080797_010 |
0.1129769339 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 16 |
Hb_000462_040 |
0.1130386502 |
- |
- |
ATSMC2, putative [Ricinus communis] |
| 17 |
Hb_000230_490 |
0.114862211 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000176_050 |
0.1157163273 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 19 |
Hb_000000_290 |
0.1173491154 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 20 |
Hb_001976_100 |
0.1248715501 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |