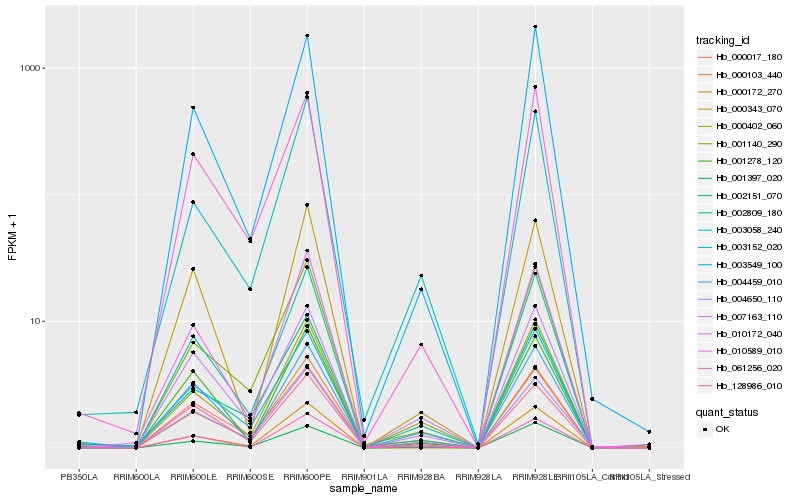
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002809_180 |
0.0 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 2 |
Hb_010172_040 |
0.0521312569 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001397_020 |
0.0613381582 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 4 |
Hb_001140_290 |
0.0651904834 |
- |
- |
hypothetical protein JCGZ_26107 [Jatropha curcas] |
| 5 |
Hb_003549_100 |
0.0677277605 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 6 |
Hb_010589_010 |
0.0694615681 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 7 |
Hb_003152_020 |
0.0705874602 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 8 |
Hb_004650_110 |
0.0713234199 |
- |
- |
hypothetical protein POPTR_0002s08760g [Populus trichocarpa] |
| 9 |
Hb_000103_440 |
0.0727132072 |
- |
- |
hypothetical protein POPTR_0008s15900g [Populus trichocarpa] |
| 10 |
Hb_061256_020 |
0.0728703417 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 11 |
Hb_000017_180 |
0.0748928883 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 12 |
Hb_001278_120 |
0.0756501507 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 13 |
Hb_000172_270 |
0.075949377 |
- |
- |
hypothetical protein JCGZ_24087 [Jatropha curcas] |
| 14 |
Hb_004459_010 |
0.0787635473 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 15 |
Hb_000343_070 |
0.0816560545 |
- |
- |
hypothetical protein JCGZ_06265 [Jatropha curcas] |
| 16 |
Hb_003058_240 |
0.0829920175 |
- |
- |
PREDICTED: tubulin beta-5 chain-like [Jatropha curcas] |
| 17 |
Hb_007163_110 |
0.0833499332 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 18 |
Hb_000402_060 |
0.0857215618 |
- |
- |
PREDICTED: uncharacterized protein LOC105644642 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002151_070 |
0.0880559771 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 2-like [Jatropha curcas] |
| 20 |
Hb_128986_010 |
0.0890537703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |