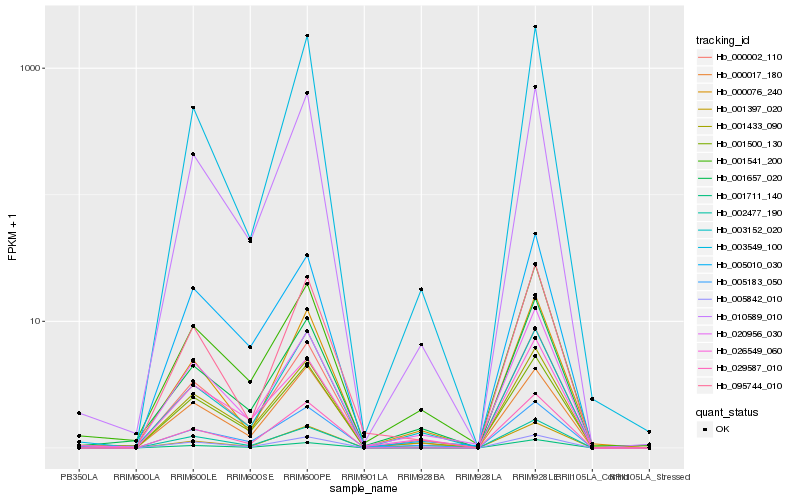
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000002_110 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_B02140 [Eucalyptus grandis] |
| 2 |
Hb_001433_090 |
0.0575756746 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 3 |
Hb_010589_010 |
0.0662606309 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 4 |
Hb_001500_130 |
0.067618145 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 5 |
Hb_001397_020 |
0.0691523838 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 6 |
Hb_005183_050 |
0.0750046644 |
- |
- |
hypothetical protein RCOM_1590900 [Ricinus communis] |
| 7 |
Hb_020956_030 |
0.0819429179 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 8 |
Hb_001657_020 |
0.0844811777 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 9 |
Hb_002477_190 |
0.0849201568 |
- |
- |
hypothetical protein EUGRSUZ_C03380, partial [Eucalyptus grandis] |
| 10 |
Hb_003152_020 |
0.0856864612 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 11 |
Hb_001541_200 |
0.0874288509 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 12 |
Hb_029587_010 |
0.0877554821 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase [Glycine soja] |
| 13 |
Hb_005842_010 |
0.0888051082 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 14 |
Hb_000017_180 |
0.0889965582 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 15 |
Hb_001711_140 |
0.0897467587 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 16 |
Hb_026549_060 |
0.0900799165 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 17 |
Hb_000076_240 |
0.0913066494 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 18 |
Hb_005010_030 |
0.0920466487 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |
| 19 |
Hb_003549_100 |
0.0924239898 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 20 |
Hb_095744_010 |
0.0941556095 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Fragaria vesca subsp. vesca] |