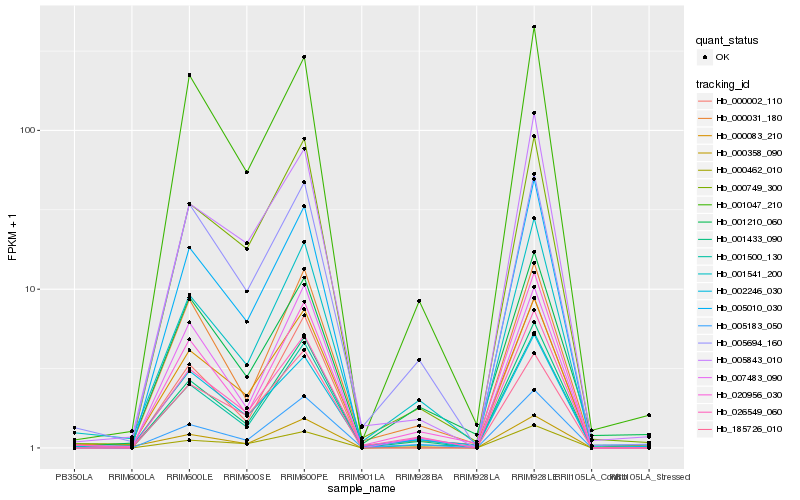
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000083_210 |
0.0 |
- |
- |
Major facilitator superfamily protein, putative [Theobroma cacao] |
| 2 |
Hb_000749_300 |
0.0690317975 |
- |
- |
PREDICTED: uncharacterized protein LOC105129180 [Populus euphratica] |
| 3 |
Hb_001500_130 |
0.0809487566 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 4 |
Hb_005010_030 |
0.0852638313 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |
| 5 |
Hb_002246_030 |
0.0891280056 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 6 |
Hb_001047_210 |
0.0895043251 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 7 |
Hb_001433_090 |
0.0944383561 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 8 |
Hb_001541_200 |
0.0949220554 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 9 |
Hb_026549_060 |
0.0955867912 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 10 |
Hb_005183_050 |
0.0969166429 |
- |
- |
hypothetical protein RCOM_1590900 [Ricinus communis] |
| 11 |
Hb_000031_180 |
0.0970488882 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 12 |
Hb_005843_010 |
0.09727496 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 13 |
Hb_000002_110 |
0.1016850154 |
- |
- |
hypothetical protein EUGRSUZ_B02140 [Eucalyptus grandis] |
| 14 |
Hb_000462_010 |
0.1017362227 |
- |
- |
hypothetical protein VITISV_041696 [Vitis vinifera] |
| 15 |
Hb_185726_010 |
0.1028553561 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 16 |
Hb_000358_090 |
0.1036663631 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_001210_060 |
0.1063386167 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 18 |
Hb_020956_030 |
0.1065626432 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 19 |
Hb_005694_160 |
0.1066520457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007483_090 |
0.1070937657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |