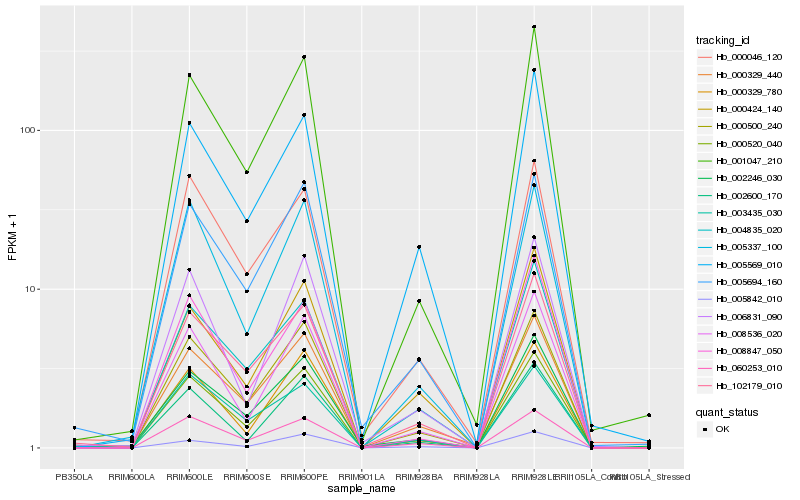
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000520_040 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 2 |
Hb_000500_240 |
0.0450979183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001047_210 |
0.0574666072 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 4 |
Hb_000329_440 |
0.0580573745 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 5 |
Hb_008536_020 |
0.0595536791 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 6 |
Hb_005337_100 |
0.0624107043 |
- |
- |
PREDICTED: uncharacterized protein LOC105629349 [Jatropha curcas] |
| 7 |
Hb_060253_010 |
0.0676201426 |
- |
- |
hypothetical protein JCGZ_14707 [Jatropha curcas] |
| 8 |
Hb_005694_160 |
0.0706566699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_102179_010 |
0.0708689305 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_000329_780 |
0.0729008904 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 11 |
Hb_004835_020 |
0.0767319146 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 12 |
Hb_000046_120 |
0.0833792422 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 13 |
Hb_003435_030 |
0.0847951041 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_005569_010 |
0.0850712885 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 15 |
Hb_002600_170 |
0.0864314144 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002246_030 |
0.0865017034 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 17 |
Hb_000424_140 |
0.0866078355 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_008847_050 |
0.0866551722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_006831_090 |
0.0867100499 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005842_010 |
0.0888794024 |
- |
- |
nitrate transporter, putative [Ricinus communis] |