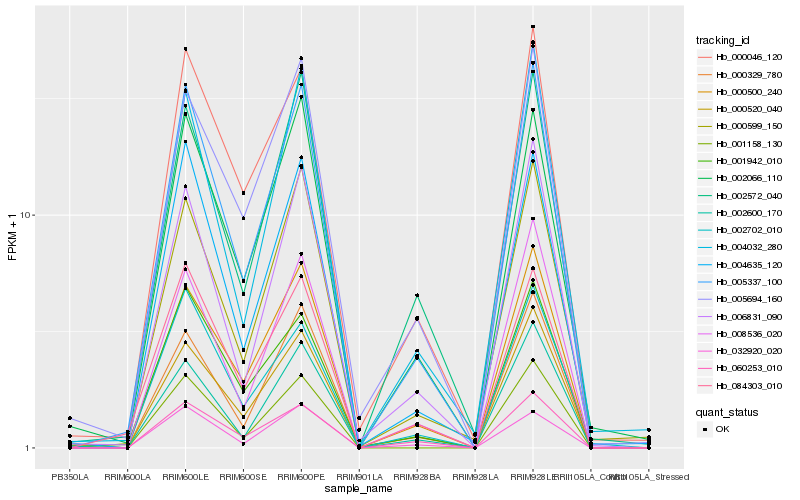
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005337_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629349 [Jatropha curcas] |
| 2 |
Hb_000520_040 |
0.0624107043 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 3 |
Hb_008536_020 |
0.0732226036 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 4 |
Hb_000500_240 |
0.0778670948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000329_780 |
0.0787956731 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 6 |
Hb_084303_010 |
0.0788940237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004635_120 |
0.0792919167 |
- |
- |
PREDICTED: rho GTPase-activating protein 4-like [Jatropha curcas] |
| 8 |
Hb_006831_090 |
0.0795762395 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 9 |
Hb_060253_010 |
0.0797623274 |
- |
- |
hypothetical protein JCGZ_14707 [Jatropha curcas] |
| 10 |
Hb_000599_150 |
0.0810913012 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |
| 11 |
Hb_004032_280 |
0.0840865989 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 12 |
Hb_001158_130 |
0.0846178928 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 13 |
Hb_000046_120 |
0.0853955977 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 14 |
Hb_002066_110 |
0.087895692 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 15 |
Hb_002572_040 |
0.0881510797 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 16 |
Hb_001942_010 |
0.0895105202 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 17 |
Hb_002702_010 |
0.0915032906 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 18 |
Hb_002600_170 |
0.0916573365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_032920_020 |
0.0930718087 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 20 |
Hb_005694_160 |
0.0937192178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |