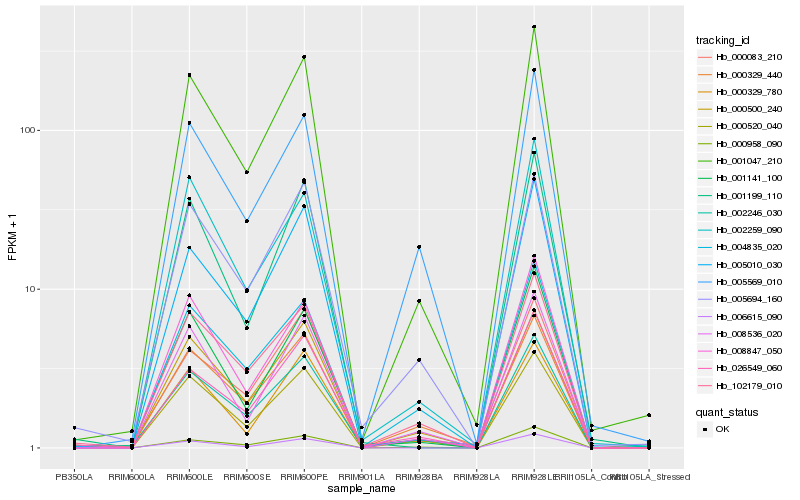
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001047_210 |
0.0 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 2 |
Hb_000520_040 |
0.0574666072 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 3 |
Hb_102179_010 |
0.0698733961 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_002246_030 |
0.0711190596 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_000958_090 |
0.0715568303 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 6 |
Hb_008847_050 |
0.0727009523 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000500_240 |
0.072710948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005010_030 |
0.0739979632 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |
| 9 |
Hb_000329_440 |
0.0777037404 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 10 |
Hb_006615_090 |
0.0778479791 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 11 |
Hb_004835_020 |
0.0785866777 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 12 |
Hb_008536_020 |
0.07882637 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 13 |
Hb_002259_090 |
0.0812585793 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 14 |
Hb_001141_100 |
0.0821475111 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 15 |
Hb_001199_110 |
0.085225754 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 16 |
Hb_026549_060 |
0.0861468005 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 17 |
Hb_005694_160 |
0.0874502511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000083_210 |
0.0895043251 |
- |
- |
Major facilitator superfamily protein, putative [Theobroma cacao] |
| 19 |
Hb_000329_780 |
0.0916417816 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 20 |
Hb_005569_010 |
0.0925100148 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |