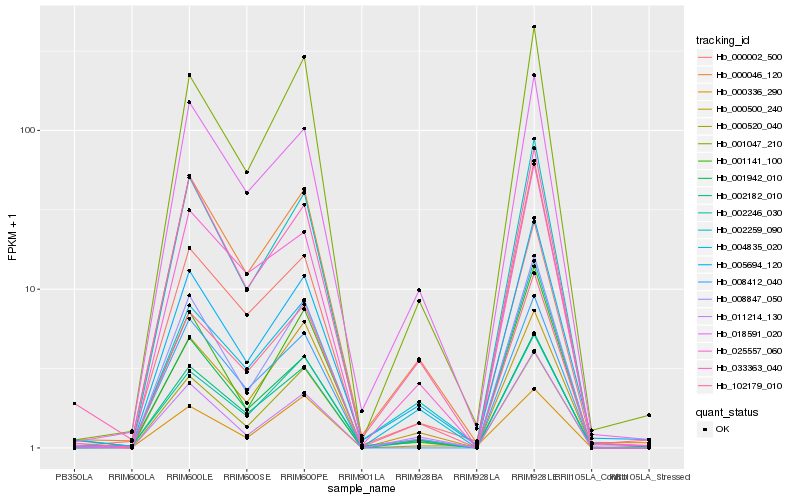
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002182_010 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase-like [Jatropha curcas] |
| 2 |
Hb_102179_010 |
0.0670035556 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_033363_040 |
0.0818200372 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 4 |
Hb_002246_030 |
0.0874641581 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_004835_020 |
0.0903275477 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 6 |
Hb_001047_210 |
0.0994235302 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 7 |
Hb_000500_240 |
0.102599115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000002_500 |
0.1028306218 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X1 [Jatropha curcas] |
| 9 |
Hb_008412_040 |
0.1032867939 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_002259_090 |
0.1044244147 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 11 |
Hb_005694_120 |
0.1053017381 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 12 |
Hb_018591_020 |
0.1062248752 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_000046_120 |
0.1065723574 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 14 |
Hb_008847_050 |
0.1072786948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000520_040 |
0.1094350163 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 16 |
Hb_011214_130 |
0.1095451309 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 [Jatropha curcas] |
| 17 |
Hb_001942_010 |
0.1102233446 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 18 |
Hb_025557_060 |
0.1121287473 |
transcription factor |
TF Family: MYB |
Myb-related protein 306, putative isoform 1 [Theobroma cacao] |
| 19 |
Hb_001141_100 |
0.1122810376 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 20 |
Hb_000336_290 |
0.112915837 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |