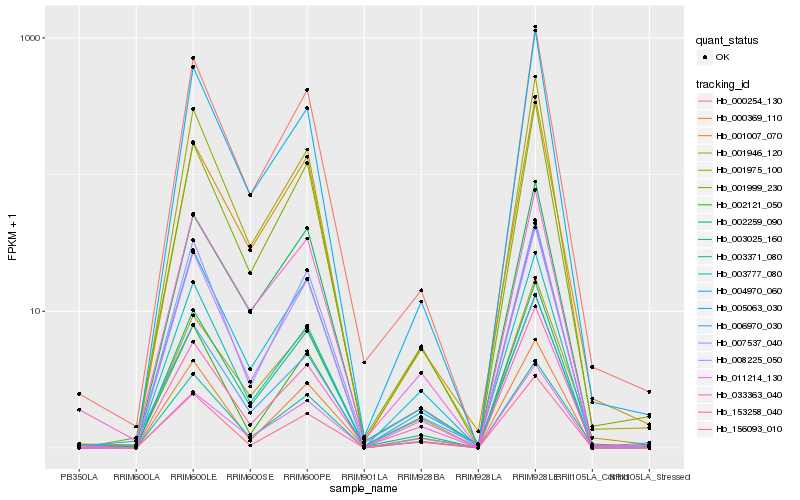
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004970_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100259061 [Vitis vinifera] |
| 2 |
Hb_003025_160 |
0.0514669728 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 3 |
Hb_008225_050 |
0.0530819262 |
- |
- |
hypothetical protein PRUPE_ppa004332mg [Prunus persica] |
| 4 |
Hb_007537_040 |
0.0594743211 |
- |
- |
PREDICTED: uncharacterized protein LOC105803495 [Gossypium raimondii] |
| 5 |
Hb_000254_130 |
0.0621095615 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_001999_230 |
0.0653230038 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 7 |
Hb_153258_040 |
0.0668236773 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 8 |
Hb_033363_040 |
0.0695266237 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 9 |
Hb_003777_080 |
0.0698640127 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 10 |
Hb_011214_130 |
0.0699400152 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 [Jatropha curcas] |
| 11 |
Hb_003371_080 |
0.0715059201 |
- |
- |
hypothetical protein POPTR_0001s23660g [Populus trichocarpa] |
| 12 |
Hb_001975_100 |
0.0734045965 |
- |
- |
chlorophyll a-b binding protein 151, chloroplastic [Jatropha curcas] |
| 13 |
Hb_156093_010 |
0.0763058596 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 14 |
Hb_005063_030 |
0.0778494039 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000369_110 |
0.0783742587 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_002259_090 |
0.0790763698 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 17 |
Hb_002121_050 |
0.079370528 |
- |
- |
hypothetical protein EUGRSUZ_D02586 [Eucalyptus grandis] |
| 18 |
Hb_001946_120 |
0.0797706003 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 19 |
Hb_001007_070 |
0.0813466984 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 20 |
Hb_006970_030 |
0.0830450561 |
- |
- |
synaptotagmin, putative [Ricinus communis] |