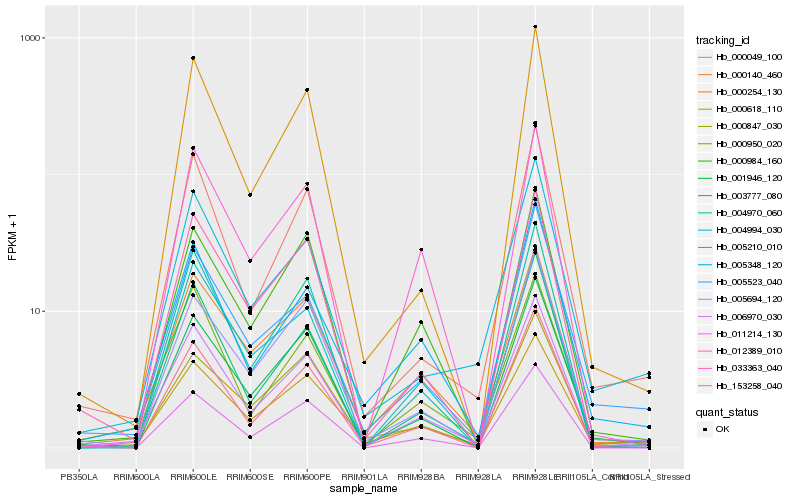
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006970_030 |
0.0 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 2 |
Hb_153258_040 |
0.0715128436 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 3 |
Hb_003777_080 |
0.0779272922 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 4 |
Hb_000140_460 |
0.0780087122 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 5 |
Hb_004970_060 |
0.0830450561 |
- |
- |
PREDICTED: uncharacterized protein LOC100259061 [Vitis vinifera] |
| 6 |
Hb_000950_020 |
0.086297893 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase [NAD(+)] GPDHC1, cytosolic [Jatropha curcas] |
| 7 |
Hb_000049_100 |
0.0883632603 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 8 |
Hb_001946_120 |
0.0932049011 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 9 |
Hb_005348_120 |
0.0960630673 |
- |
- |
PREDICTED: chlorophyll a-b binding protein, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000984_160 |
0.0960644864 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_012389_010 |
0.0984355806 |
- |
- |
hypothetical protein POPTR_0018s11820g [Populus trichocarpa] |
| 12 |
Hb_005694_120 |
0.0984564209 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 13 |
Hb_005523_040 |
0.098801895 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 14 |
Hb_000618_110 |
0.1000174387 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 15 |
Hb_033363_040 |
0.1006147472 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 16 |
Hb_011214_130 |
0.1011750623 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 [Jatropha curcas] |
| 17 |
Hb_000254_130 |
0.1018803373 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000847_030 |
0.1029056657 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 19 |
Hb_005210_010 |
0.1036472186 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 20 |
Hb_004994_030 |
0.1047213437 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |