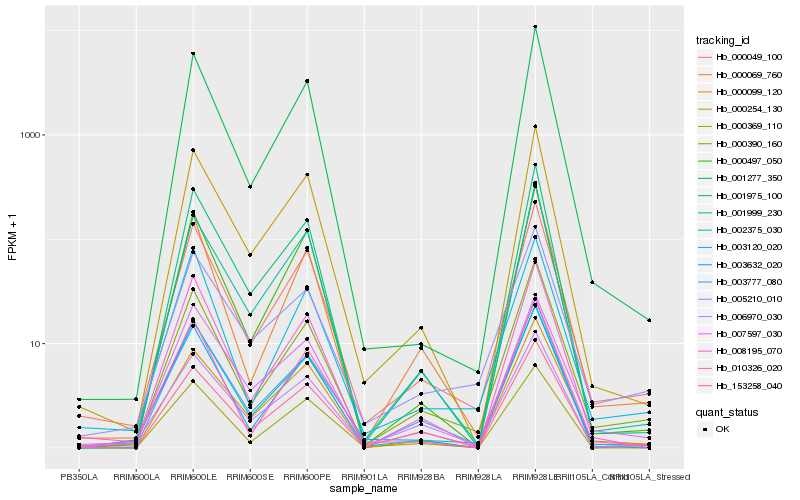
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000049_100 |
0.0 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 2 |
Hb_000390_160 |
0.0692077222 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 3 |
Hb_000254_130 |
0.070423475 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_003120_020 |
0.0743765728 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 13-like [Prunus mume] |
| 5 |
Hb_002375_030 |
0.074974947 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 6 |
Hb_003632_020 |
0.0782250242 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 7 |
Hb_010326_020 |
0.0827088593 |
- |
- |
PREDICTED: thioredoxin-like 4, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003777_080 |
0.0839151208 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 9 |
Hb_007597_030 |
0.084802739 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_008195_070 |
0.0852679198 |
- |
- |
PREDICTED: thylakoid lumenal protein At1g12250, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_001999_230 |
0.0859895939 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 12 |
Hb_153258_040 |
0.0870302044 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 13 |
Hb_000369_110 |
0.0870359846 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_001277_350 |
0.0872076907 |
- |
- |
early light-induced protein, putative [Ricinus communis] |
| 15 |
Hb_000069_760 |
0.0872369076 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 16 |
Hb_006970_030 |
0.0883632603 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 17 |
Hb_005210_010 |
0.0884483122 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 18 |
Hb_000099_120 |
0.0887698665 |
- |
- |
PREDICTED: GDT1-like protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 19 |
Hb_001975_100 |
0.0890445085 |
- |
- |
chlorophyll a-b binding protein 151, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000497_050 |
0.0890954473 |
- |
- |
PREDICTED: photosystem II repair protein PSB27-H1, chloroplastic [Jatropha curcas] |