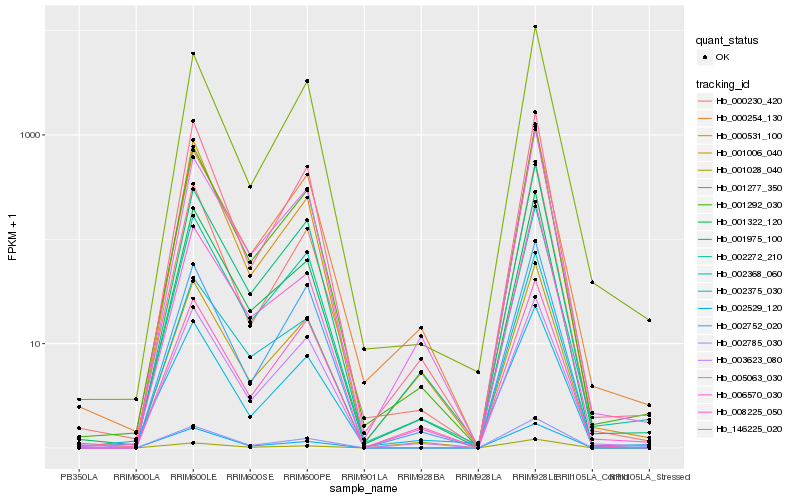
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001006_040 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g13770, chloroplastic-like isoform X1 [Populus euphratica] |
| 2 |
Hb_002785_030 |
0.0324693433 |
- |
- |
Polyketide cyclase/dehydrase and lipid transport superfamily protein [Theobroma cacao] |
| 3 |
Hb_001292_030 |
0.0347112515 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001975_100 |
0.0467242527 |
- |
- |
chlorophyll a-b binding protein 151, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001322_120 |
0.0478491187 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 6 |
Hb_005063_030 |
0.0579385106 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_001028_040 |
0.0602011399 |
- |
- |
hypothetical protein POPTR_0018s09860g [Populus trichocarpa] |
| 8 |
Hb_006570_030 |
0.0606865056 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 9 |
Hb_146225_020 |
0.0610493161 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 10 |
Hb_002272_210 |
0.0613173504 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002529_120 |
0.062287445 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7-like [Jatropha curcas] |
| 12 |
Hb_003623_080 |
0.0642045977 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Populus euphratica] |
| 13 |
Hb_000230_420 |
0.0648586784 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 14 |
Hb_002368_060 |
0.0657869002 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000531_100 |
0.0659976228 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_002752_020 |
0.0670646628 |
- |
- |
FKBP-type peptidyl-prolyl cis-trans isomerase 3 family protein [Populus trichocarpa] |
| 17 |
Hb_001277_350 |
0.0694778964 |
- |
- |
early light-induced protein, putative [Ricinus communis] |
| 18 |
Hb_002375_030 |
0.0695009772 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 19 |
Hb_000254_130 |
0.0696131633 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_008225_050 |
0.0711071228 |
- |
- |
hypothetical protein PRUPE_ppa004332mg [Prunus persica] |