| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
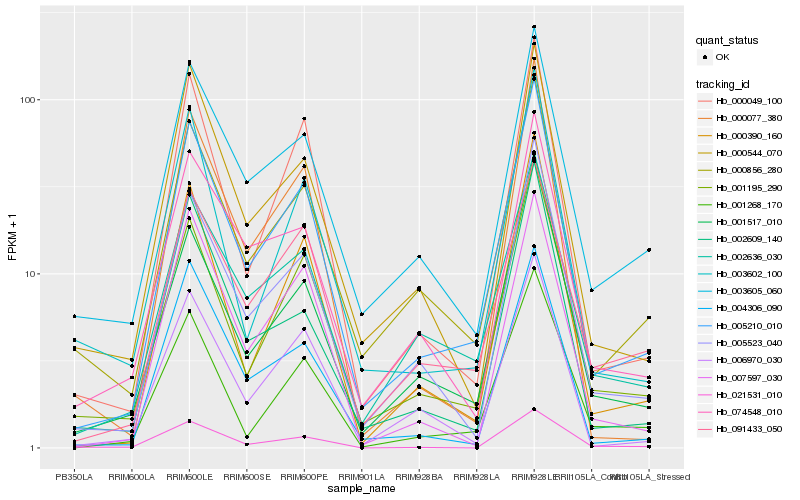
Hb_005210_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 2 |
Hb_005523_040 |
0.0833899089 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 3 |
Hb_000049_100 |
0.0884483122 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 4 |
Hb_000390_160 |
0.0895850534 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 5 |
Hb_001195_290 |
0.0896214837 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 6 |
Hb_000856_280 |
0.0914598059 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 7 |
Hb_006970_030 |
0.1036472186 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 8 |
Hb_001517_010 |
0.1037118731 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002636_030 |
0.1057086669 |
- |
- |
hypothetical protein JCGZ_11800 [Jatropha curcas] |
| 10 |
Hb_000544_070 |
0.1068115675 |
- |
- |
Peptidyl-prolyl cis-trans isomerase FKBP17-2 [Morus notabilis] |
| 11 |
Hb_004306_090 |
0.1068140896 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001268_170 |
0.1071007345 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 13 |
Hb_007597_030 |
0.1090189911 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_003602_100 |
0.109301787 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_091433_050 |
0.110034914 |
- |
- |
PREDICTED: uncharacterized protein LOC105646626 [Jatropha curcas] |
| 16 |
Hb_003605_060 |
0.1100962306 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000077_380 |
0.1109934666 |
- |
- |
PREDICTED: protochlorophyllide reductase [Jatropha curcas] |
| 18 |
Hb_074548_010 |
0.112743403 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_002609_140 |
0.1135731633 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_021531_010 |
0.1137351888 |
- |
- |
receptor-kinase, putative [Ricinus communis] |