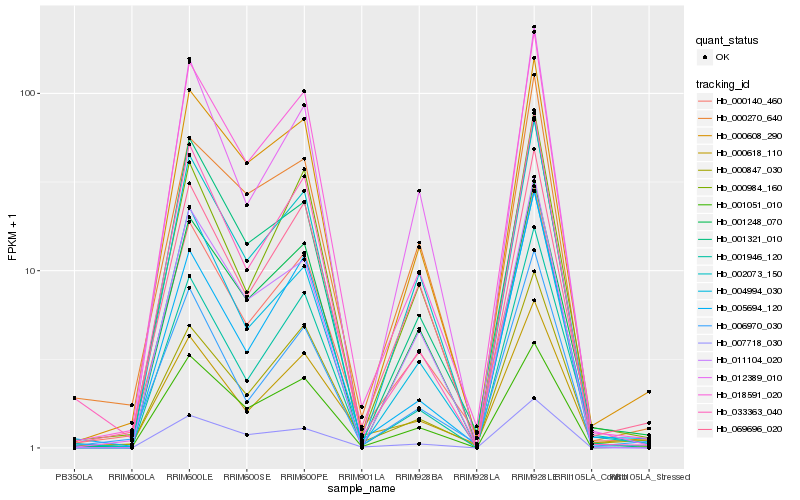
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_460 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 2 |
Hb_069696_020 |
0.0757314428 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_002073_150 |
0.0775846501 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 4 |
Hb_006970_030 |
0.0780087122 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 5 |
Hb_000618_110 |
0.0813035698 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 6 |
Hb_000984_160 |
0.0817714853 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_018591_020 |
0.0824625286 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_001051_010 |
0.0835746606 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 9 |
Hb_000847_030 |
0.0847468052 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 10 |
Hb_000608_290 |
0.0856001539 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 11 |
Hb_001248_070 |
0.0892502142 |
- |
- |
PREDICTED: probable polyamine transporter At3g19553 [Jatropha curcas] |
| 12 |
Hb_012389_010 |
0.0895189075 |
- |
- |
hypothetical protein POPTR_0018s11820g [Populus trichocarpa] |
| 13 |
Hb_004994_030 |
0.0923689305 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 14 |
Hb_011104_020 |
0.0931850892 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 15 |
Hb_001321_010 |
0.0943596467 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 16 |
Hb_033363_040 |
0.0962800436 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 17 |
Hb_007718_030 |
0.0983528346 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 18 |
Hb_000270_640 |
0.0992730112 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 19 |
Hb_005694_120 |
0.0998562782 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 20 |
Hb_001946_120 |
0.0999782094 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |