| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
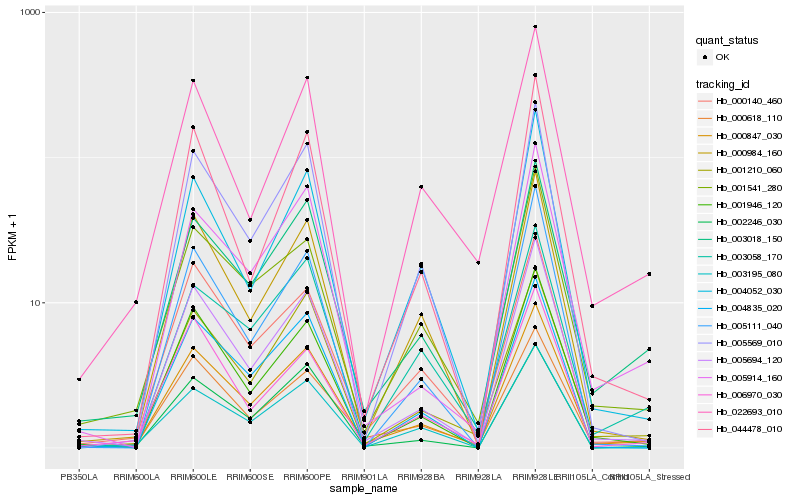
Hb_000847_030 |
0.0 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 2 |
Hb_005694_120 |
0.0557929248 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 3 |
Hb_003018_150 |
0.0707106213 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 4 |
Hb_000618_110 |
0.0789356431 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 5 |
Hb_000984_160 |
0.079111447 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001946_120 |
0.0799595585 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 7 |
Hb_001210_060 |
0.0818759778 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 8 |
Hb_001541_280 |
0.0844630769 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 9 |
Hb_000140_460 |
0.0847468052 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 10 |
Hb_005569_010 |
0.0865867326 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 11 |
Hb_004835_020 |
0.0906961619 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 12 |
Hb_004052_030 |
0.0929094936 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 13 |
Hb_002246_030 |
0.0970013435 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 14 |
Hb_005914_160 |
0.0971018289 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003195_080 |
0.0982129649 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 16 |
Hb_006970_030 |
0.1029056657 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 17 |
Hb_003058_170 |
0.1031113526 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 18 |
Hb_044478_010 |
0.1039757871 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 19 |
Hb_022693_010 |
0.1040269614 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 20 |
Hb_005111_040 |
0.1056245248 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |