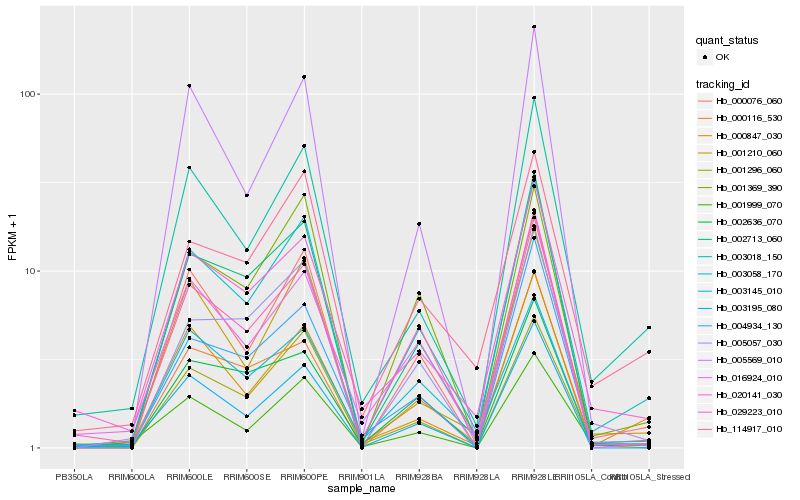
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003058_170 |
0.0 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000076_060 |
0.0795964348 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 3 |
Hb_003018_150 |
0.0860654958 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 4 |
Hb_002713_060 |
0.0940873611 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 5 |
Hb_001210_060 |
0.1024193843 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 6 |
Hb_000847_030 |
0.1031113526 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 7 |
Hb_029223_010 |
0.1044063634 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 8 |
Hb_001369_390 |
0.105173077 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 9 |
Hb_020141_030 |
0.1106183478 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 10 |
Hb_001296_060 |
0.1126137293 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 11 |
Hb_000116_530 |
0.1126477563 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 3; Short=HMG-CoA reductase 3 [Hevea brasiliensis] |
| 12 |
Hb_114917_010 |
0.1126877054 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003145_010 |
0.1139335125 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 14 |
Hb_002636_070 |
0.1147449487 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 15 |
Hb_005569_010 |
0.1162742766 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 16 |
Hb_005057_030 |
0.1164120342 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 17 |
Hb_004934_130 |
0.1191130487 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 18 |
Hb_003195_080 |
0.1192133178 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 19 |
Hb_001999_070 |
0.119987025 |
- |
- |
PREDICTED: ammonium transporter 1 member 3 [Jatropha curcas] |
| 20 |
Hb_016924_010 |
0.1205247377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |