| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
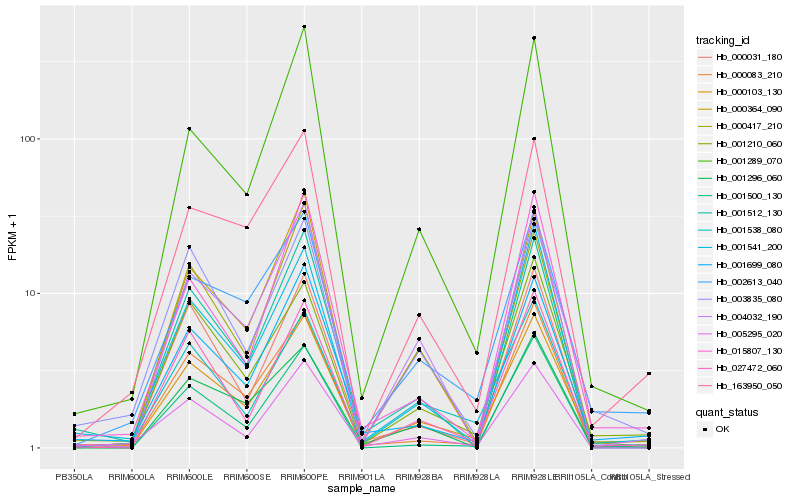
Hb_000103_130 |
0.0 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 2 |
Hb_002613_040 |
0.0761824555 |
- |
- |
phosphate translocator-related family protein [Populus trichocarpa] |
| 3 |
Hb_001699_080 |
0.083249835 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_001538_080 |
0.0889978792 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001210_060 |
0.0948638386 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 6 |
Hb_163950_050 |
0.0987820007 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_004032_190 |
0.0993261444 |
- |
- |
PREDICTED: probable cellulose synthase A catalytic subunit 3 [UDP-forming] isoform X1 [Jatropha curcas] |
| 8 |
Hb_000364_090 |
0.1060106592 |
- |
- |
PREDICTED: filament-like plant protein 7 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003835_080 |
0.1063123804 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001541_200 |
0.1083637009 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 11 |
Hb_027472_060 |
0.1088591543 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 12 |
Hb_005295_020 |
0.1093527329 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 13 |
Hb_000083_210 |
0.1098143871 |
- |
- |
Major facilitator superfamily protein, putative [Theobroma cacao] |
| 14 |
Hb_001512_130 |
0.1110793767 |
- |
- |
PREDICTED: uncharacterized protein LOC105633633 [Jatropha curcas] |
| 15 |
Hb_001289_070 |
0.1135686144 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 16 |
Hb_000031_180 |
0.1154659192 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 17 |
Hb_000417_210 |
0.1155439064 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_001500_130 |
0.1198909137 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 19 |
Hb_015807_130 |
0.1204117018 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001296_060 |
0.1204674843 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |