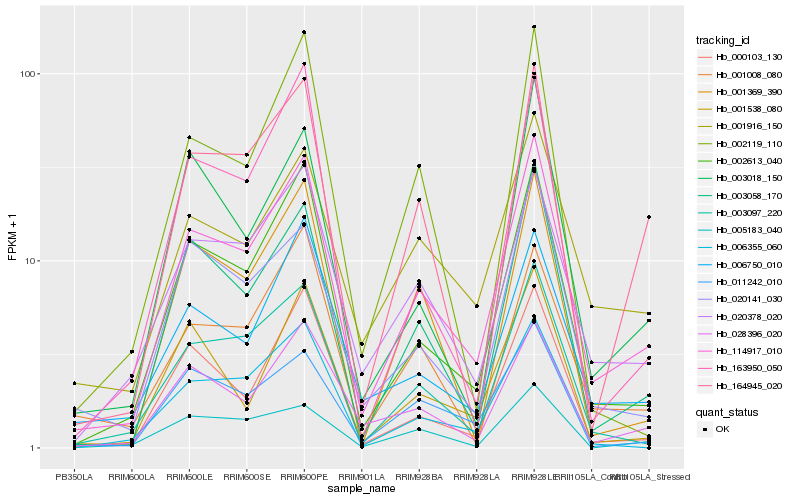
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114917_010 |
0.0 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002613_040 |
0.0890293457 |
- |
- |
phosphate translocator-related family protein [Populus trichocarpa] |
| 3 |
Hb_003097_220 |
0.1116141071 |
- |
- |
hypothetical protein POPTR_0001s06150g [Populus trichocarpa] |
| 4 |
Hb_003058_170 |
0.1126877054 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 5 |
Hb_020378_020 |
0.1147366927 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_028396_020 |
0.1200151945 |
- |
- |
PREDICTED: ras-related protein Rab7 [Jatropha curcas] |
| 7 |
Hb_001916_150 |
0.1215146271 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3 [Vitis vinifera] |
| 8 |
Hb_000103_130 |
0.1242225177 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 9 |
Hb_006750_010 |
0.1265993541 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 10 |
Hb_011242_010 |
0.1314914553 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 11 |
Hb_164945_020 |
0.1325845633 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 12 |
Hb_006355_060 |
0.1334361254 |
- |
- |
PREDICTED: transcription factor PIF1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_020141_030 |
0.1358262591 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 14 |
Hb_001369_390 |
0.1368177595 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 15 |
Hb_003018_150 |
0.1409510532 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 16 |
Hb_163950_050 |
0.1438961952 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_001008_080 |
0.1441328286 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 18 |
Hb_001538_080 |
0.1454019772 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005183_040 |
0.1476102261 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 20 |
Hb_002119_110 |
0.148870084 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |