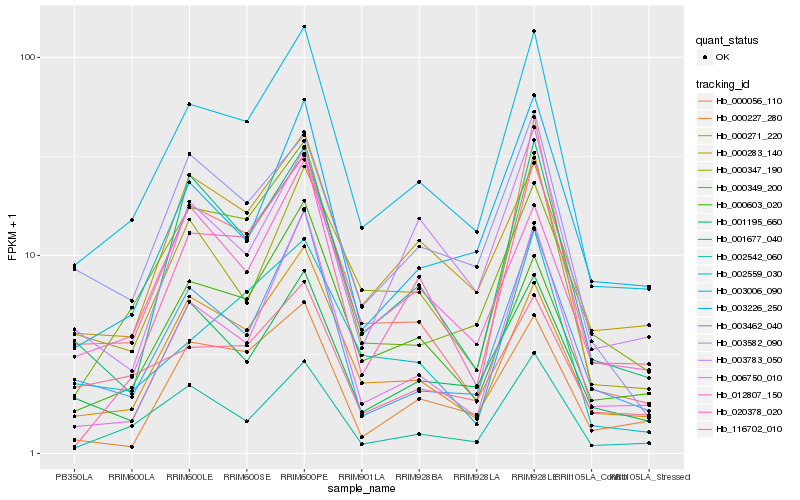
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003006_090 |
0.0 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 2 |
Hb_000271_220 |
0.1084620029 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 3 |
Hb_002542_060 |
0.1122415553 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 4 |
Hb_003462_040 |
0.1141164927 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_001195_660 |
0.1183702931 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 6 |
Hb_001677_040 |
0.118818321 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000603_020 |
0.1193779228 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 8 |
Hb_020378_020 |
0.1205594077 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000347_190 |
0.1233106865 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 10 |
Hb_006750_010 |
0.1241681274 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 11 |
Hb_000349_200 |
0.1256375501 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g12500 [Jatropha curcas] |
| 12 |
Hb_000283_140 |
0.1261039093 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000056_110 |
0.1264619685 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 14 |
Hb_012807_150 |
0.1274020133 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 15 |
Hb_003226_250 |
0.1279287291 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 16 |
Hb_003582_090 |
0.1291990608 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 6-like [Jatropha curcas] |
| 17 |
Hb_002559_030 |
0.1313667652 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 18 |
Hb_116702_010 |
0.1349524273 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 19 |
Hb_000227_280 |
0.1375317097 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 20 |
Hb_003783_050 |
0.1389312491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |