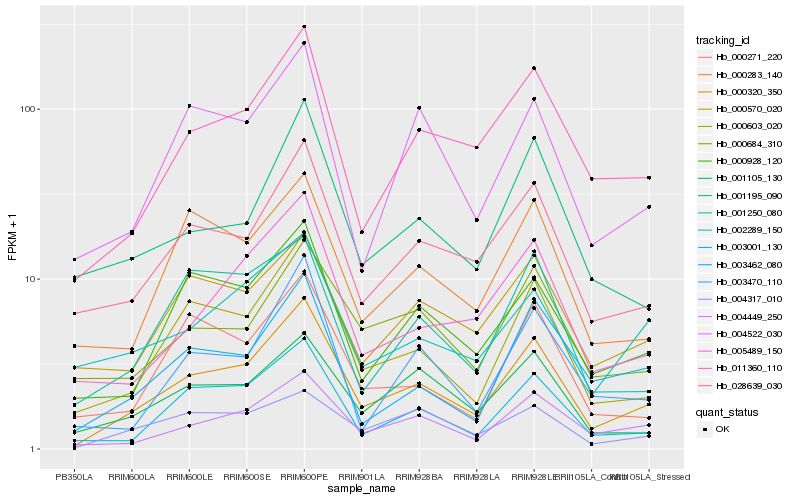
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000320_350 |
0.0 |
- |
- |
hypothetical protein RCOM_0904330 [Ricinus communis] |
| 2 |
Hb_000603_020 |
0.1099572835 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004449_250 |
0.1194832469 |
- |
- |
PREDICTED: protein EI24 homolog [Jatropha curcas] |
| 4 |
Hb_011360_110 |
0.122929726 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 5 |
Hb_003001_130 |
0.1257581114 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_028639_030 |
0.1284706191 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 7 |
Hb_000928_120 |
0.136816338 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 8 |
Hb_005489_150 |
0.1422839934 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 9 |
Hb_004522_030 |
0.1480454203 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 10 |
Hb_001105_130 |
0.1501709294 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 11 |
Hb_001250_080 |
0.1509251636 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g23390 [Jatropha curcas] |
| 12 |
Hb_000283_140 |
0.1513502755 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_003462_080 |
0.1521150842 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_000684_310 |
0.152739203 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 15 |
Hb_004317_010 |
0.152802998 |
- |
- |
hypothetical protein CICLE_v10011038mg [Citrus clementina] |
| 16 |
Hb_003470_110 |
0.1552867552 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_001195_090 |
0.1556744732 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 18 |
Hb_000570_020 |
0.1564129706 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 19 |
Hb_002289_150 |
0.1570327267 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 20 |
Hb_000271_220 |
0.1582751722 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |