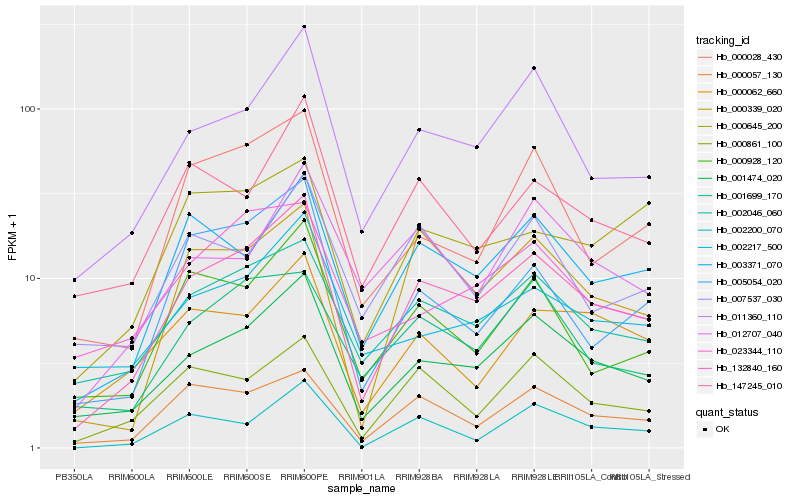
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_132840_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001474_020 |
0.0858429012 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 3 |
Hb_011360_110 |
0.1059195385 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 4 |
Hb_002046_060 |
0.1260132131 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 5 |
Hb_003371_070 |
0.1282259341 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000057_130 |
0.1322278881 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 7 |
Hb_000339_020 |
0.1396257878 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 12-like [Jatropha curcas] |
| 8 |
Hb_000861_100 |
0.1397380901 |
- |
- |
PREDICTED: probable fucosyltransferase 8 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000928_120 |
0.1399185288 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 10 |
Hb_000028_430 |
0.1446116652 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005054_020 |
0.1463181082 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 12 |
Hb_147245_010 |
0.1484362442 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 13 |
Hb_002217_500 |
0.1489151806 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 14 |
Hb_002200_070 |
0.1513489375 |
- |
- |
PREDICTED: endoglucanase 9-like [Populus euphratica] |
| 15 |
Hb_001699_170 |
0.153826977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000062_660 |
0.1556226673 |
- |
- |
hypothetical protein POPTR_0009s03470g [Populus trichocarpa] |
| 17 |
Hb_023344_110 |
0.1595171798 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 18 |
Hb_012707_040 |
0.1606216763 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 29 [Jatropha curcas] |
| 19 |
Hb_000645_200 |
0.1614536934 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 20 |
Hb_007537_030 |
0.1620115355 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |