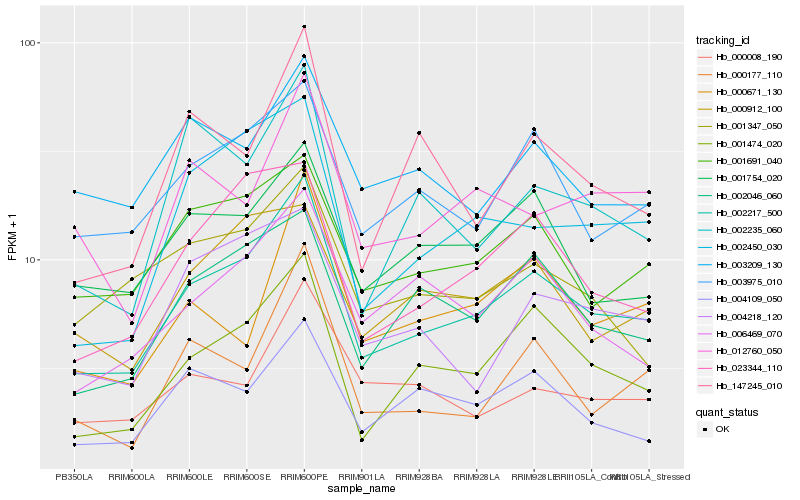
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_500 |
0.0 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 2 |
Hb_001474_020 |
0.1055966283 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 3 |
Hb_003975_010 |
0.1201365047 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 4 |
Hb_002046_060 |
0.1217942262 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 5 |
Hb_001691_040 |
0.1231289558 |
- |
- |
PREDICTED: uncharacterized protein LOC105649546 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002450_030 |
0.1245331118 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 7 |
Hb_023344_110 |
0.1262815323 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 8 |
Hb_006469_070 |
0.1286288486 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |
| 9 |
Hb_001754_020 |
0.1316703353 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 10 |
Hb_004109_050 |
0.1371789695 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |
| 11 |
Hb_000912_100 |
0.1393351 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 12 |
Hb_000008_190 |
0.139957061 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004218_120 |
0.1402838753 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_147245_010 |
0.1403757633 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 15 |
Hb_000177_110 |
0.1404601362 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 16 |
Hb_002235_060 |
0.1413246313 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 17 |
Hb_012760_050 |
0.1420725585 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 18 |
Hb_001347_050 |
0.1421626881 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 19 |
Hb_003209_130 |
0.1435294035 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 20 |
Hb_000671_130 |
0.1435997765 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |