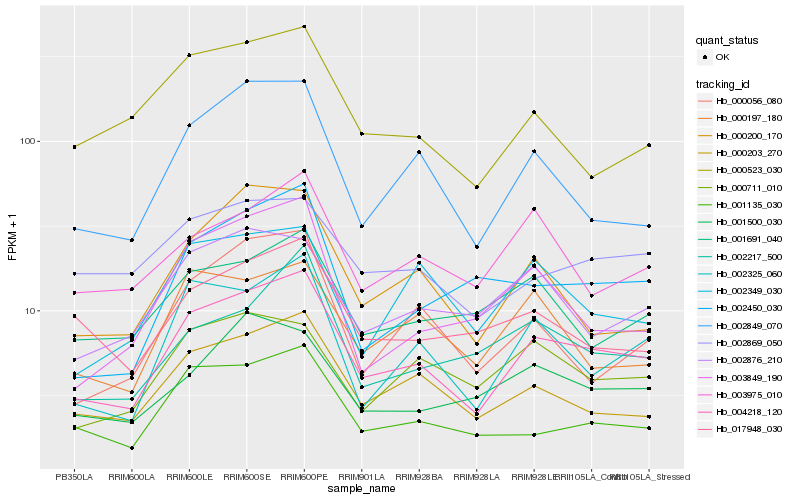
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_120 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000203_270 |
0.1094505944 |
- |
- |
alpha-xylosidase, putative [Ricinus communis] |
| 3 |
Hb_002325_060 |
0.1164148526 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 4 |
Hb_001135_030 |
0.1220611968 |
- |
- |
PREDICTED: interaptin-like [Jatropha curcas] |
| 5 |
Hb_002849_070 |
0.1248959854 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000200_170 |
0.1337592525 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 7 |
Hb_002876_210 |
0.1354928292 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 8 |
Hb_001500_030 |
0.1375166262 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002450_030 |
0.1388493568 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 10 |
Hb_002349_030 |
0.1389132952 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000056_080 |
0.13893197 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003849_190 |
0.1393963552 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 13 |
Hb_000197_180 |
0.1393997677 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 14 |
Hb_017948_030 |
0.1395699712 |
- |
- |
PREDICTED: uncharacterized protein LOC105639493 [Jatropha curcas] |
| 15 |
Hb_002869_050 |
0.1396903527 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 16 |
Hb_003975_010 |
0.139759769 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 17 |
Hb_000523_030 |
0.1398209214 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 18 |
Hb_000711_010 |
0.1399718675 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 19 |
Hb_002217_500 |
0.1402838753 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 20 |
Hb_001691_040 |
0.1402992687 |
- |
- |
PREDICTED: uncharacterized protein LOC105649546 isoform X2 [Jatropha curcas] |