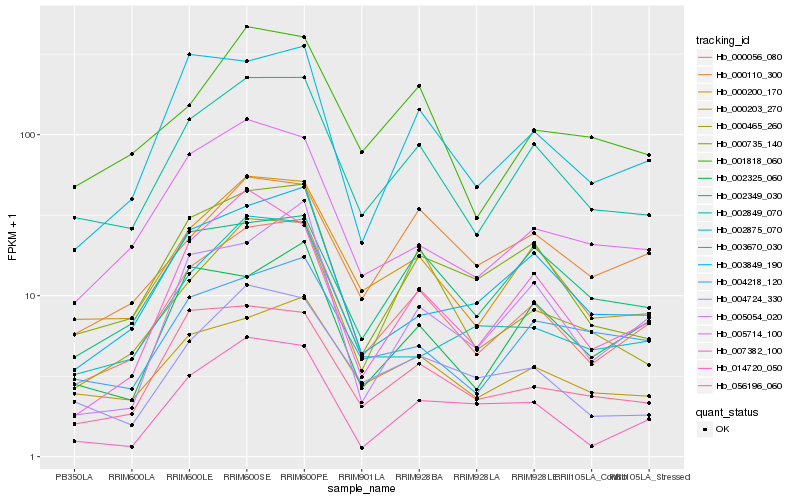
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000056_080 |
0.0 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002849_070 |
0.0759583881 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000200_170 |
0.0851062725 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 4 |
Hb_000203_270 |
0.1109356122 |
- |
- |
alpha-xylosidase, putative [Ricinus communis] |
| 5 |
Hb_003670_030 |
0.1131963698 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 6 |
Hb_000735_140 |
0.1134765444 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 7 |
Hb_003849_190 |
0.1221752085 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 8 |
Hb_001818_060 |
0.1238639999 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 5-like [Jatropha curcas] |
| 9 |
Hb_056196_060 |
0.1241845521 |
- |
- |
hexokinase [Manihot esculenta] |
| 10 |
Hb_005714_100 |
0.1270165195 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 11 |
Hb_004724_330 |
0.1277515243 |
- |
- |
Capsanthin/capsorubin synthase, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000465_260 |
0.1315136877 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 13 |
Hb_005054_020 |
0.1328873468 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 14 |
Hb_007382_100 |
0.1331887539 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 15 |
Hb_002325_060 |
0.1343573408 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 16 |
Hb_000110_300 |
0.1369983526 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 17 |
Hb_002875_070 |
0.1373464184 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 18 |
Hb_002349_030 |
0.1384981768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004218_120 |
0.13893197 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_014720_050 |
0.1389621461 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |