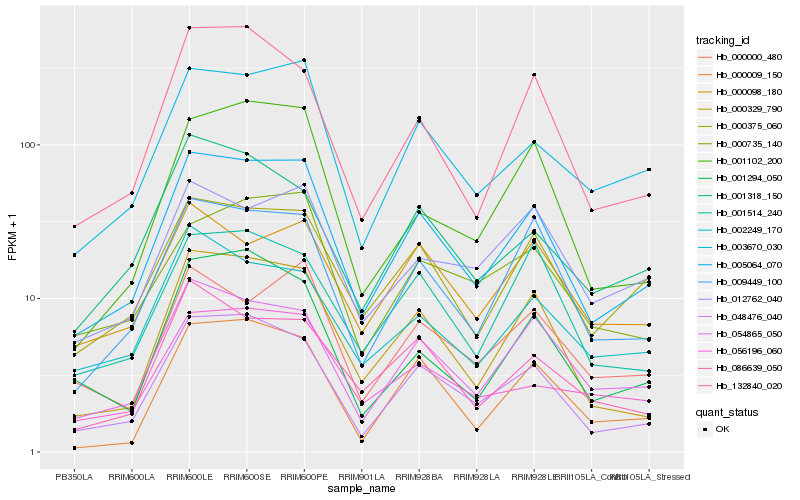
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005064_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648318 [Jatropha curcas] |
| 2 |
Hb_048476_040 |
0.0795776945 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 3 |
Hb_003670_030 |
0.0884618324 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 4 |
Hb_000329_790 |
0.0931877053 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 5 |
Hb_054865_050 |
0.0940789923 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 6 |
Hb_009449_100 |
0.0957216958 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 7 |
Hb_000375_060 |
0.1004427692 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 8 |
Hb_132840_020 |
0.1180903954 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 9 |
Hb_000009_150 |
0.1196148206 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 10 |
Hb_056196_060 |
0.1203851667 |
- |
- |
hexokinase [Manihot esculenta] |
| 11 |
Hb_086639_050 |
0.1231981752 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000000_480 |
0.1235771306 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001102_200 |
0.1238588175 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 14 |
Hb_001514_240 |
0.124704182 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 15 |
Hb_002249_170 |
0.1253420691 |
- |
- |
copine, putative [Ricinus communis] |
| 16 |
Hb_000735_140 |
0.1266398215 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 17 |
Hb_001318_150 |
0.1268364722 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 18 |
Hb_001294_050 |
0.1283584518 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 19 |
Hb_012762_040 |
0.129788649 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 20 |
Hb_000098_180 |
0.1303377706 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |