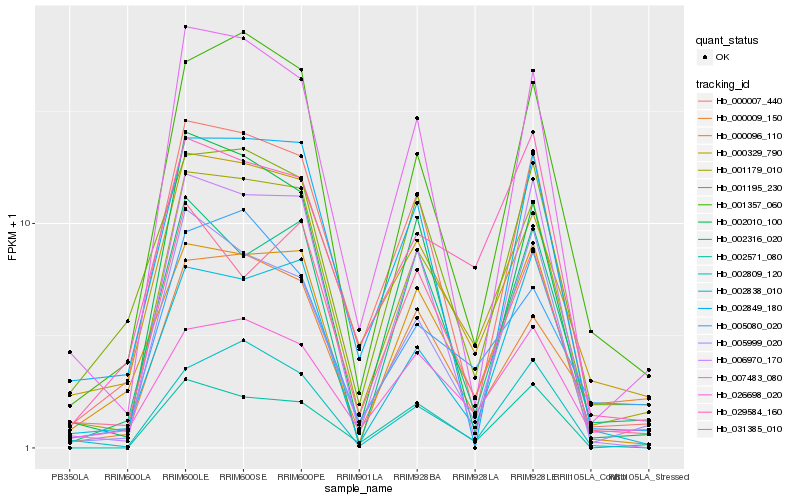
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001179_010 |
0.0 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 2 |
Hb_001357_060 |
0.1056362226 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 18-like [Cicer arietinum] |
| 3 |
Hb_000329_790 |
0.1086044589 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 4 |
Hb_000007_440 |
0.1150749467 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 5 |
Hb_007483_080 |
0.1178196775 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 6 |
Hb_000096_110 |
0.1197934555 |
- |
- |
PREDICTED: uncharacterized protein LOC105787685 isoform X1 [Gossypium raimondii] |
| 7 |
Hb_026698_020 |
0.129828814 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 8 |
Hb_001195_230 |
0.1301425505 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4 [Jatropha curcas] |
| 9 |
Hb_002838_010 |
0.1341776926 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 10 |
Hb_031385_010 |
0.1342543422 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 11 |
Hb_000009_150 |
0.1356907767 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 12 |
Hb_005999_020 |
0.1361088222 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 13 |
Hb_002809_120 |
0.1361987689 |
- |
- |
Uncharacterized protein TCM_031281 [Theobroma cacao] |
| 14 |
Hb_002010_100 |
0.1363919895 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 15 |
Hb_029584_160 |
0.1380085417 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 16 |
Hb_002571_080 |
0.1381417484 |
- |
- |
secondary cell wall-related glycosyltransferase family 14 [Populus tremula x Populus tremuloides] |
| 17 |
Hb_005080_020 |
0.139185494 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 18 |
Hb_002316_020 |
0.1397415711 |
- |
- |
PREDICTED: uncharacterized protein LOC105640407 [Jatropha curcas] |
| 19 |
Hb_006970_170 |
0.1400537546 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_002849_180 |
0.1402114591 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |