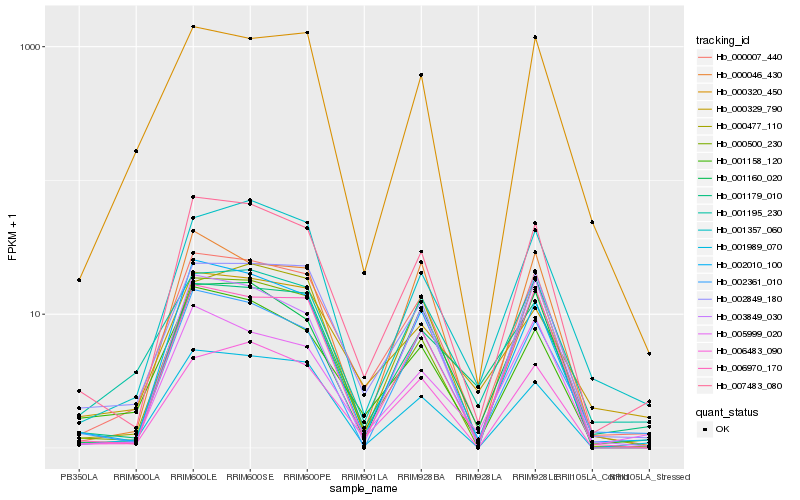
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000007_440 |
0.0 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 2 |
Hb_007483_080 |
0.0724787499 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 3 |
Hb_002849_180 |
0.0762840505 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 4 |
Hb_002010_100 |
0.1026305567 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 5 |
Hb_006483_090 |
0.1038897727 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 6 |
Hb_001158_120 |
0.1066959105 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006970_170 |
0.108196404 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_001195_230 |
0.111178839 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4 [Jatropha curcas] |
| 9 |
Hb_000320_450 |
0.1131145082 |
- |
- |
secretory peroxidase [Hevea brasiliensis] |
| 10 |
Hb_000477_110 |
0.1150413066 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 11 |
Hb_001179_010 |
0.1150749467 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 12 |
Hb_000500_230 |
0.1159539835 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 13 |
Hb_001357_060 |
0.1182918889 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 18-like [Cicer arietinum] |
| 14 |
Hb_001989_070 |
0.1185940419 |
- |
- |
PREDICTED: uncharacterized calcium-binding protein At1g02270 [Jatropha curcas] |
| 15 |
Hb_005999_020 |
0.1194326276 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 16 |
Hb_001160_020 |
0.124369158 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 17 |
Hb_002361_010 |
0.1244571319 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000329_790 |
0.1269474134 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 19 |
Hb_003849_030 |
0.1283582901 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 20 |
Hb_000046_430 |
0.1286859403 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |