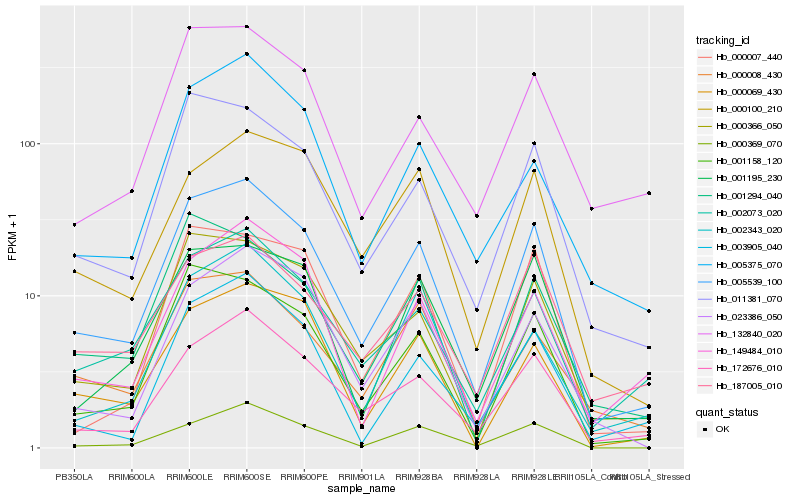
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005539_100 |
0.0 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 2 |
Hb_172676_010 |
0.0915976052 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 3 |
Hb_011381_070 |
0.1011094664 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 4 |
Hb_001158_120 |
0.1021903688 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000366_050 |
0.1029585662 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 6 |
Hb_002073_020 |
0.1075302412 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 7 |
Hb_000369_070 |
0.1207065352 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 8 |
Hb_000008_430 |
0.1226288399 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 9 |
Hb_187005_010 |
0.1234025484 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 10 |
Hb_001195_230 |
0.1237077636 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4 [Jatropha curcas] |
| 11 |
Hb_000100_210 |
0.1281711508 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |
| 12 |
Hb_023386_050 |
0.1284568135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000069_430 |
0.1288379387 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 14 |
Hb_149484_010 |
0.1309580672 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor-like [Eucalyptus grandis] |
| 15 |
Hb_132840_020 |
0.1329607661 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 16 |
Hb_000007_440 |
0.1340691091 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 17 |
Hb_001294_040 |
0.1342185778 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 18 |
Hb_005375_070 |
0.1349574885 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 19 |
Hb_003905_040 |
0.1355738389 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 20 |
Hb_002343_020 |
0.1355996619 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |