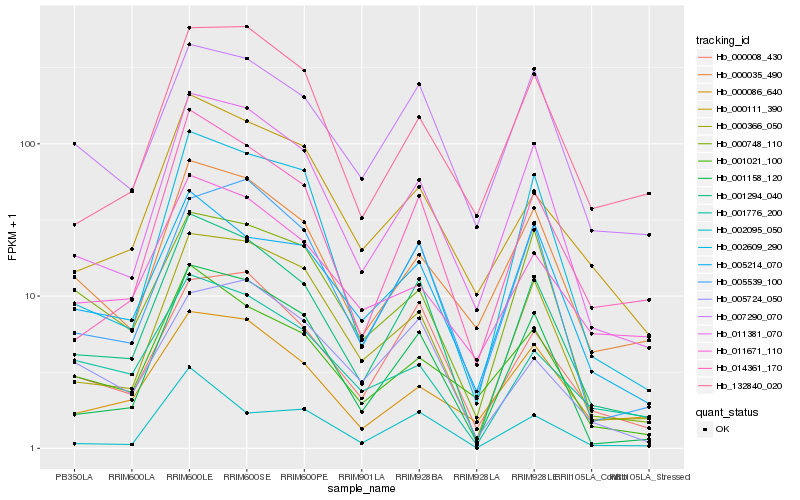
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001294_040 |
0.0 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 2 |
Hb_001158_120 |
0.0932778186 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 3 |
Hb_011381_070 |
0.0940626037 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 4 |
Hb_014361_170 |
0.1124065166 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 37 [Jatropha curcas] |
| 5 |
Hb_000366_050 |
0.1140284946 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 6 |
Hb_000008_430 |
0.1195306746 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000035_490 |
0.1340879309 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 8 |
Hb_005539_100 |
0.1342185778 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 9 |
Hb_001021_100 |
0.1395755525 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 10 |
Hb_005214_070 |
0.1397638879 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |
| 11 |
Hb_007290_070 |
0.1398125755 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 12 |
Hb_002609_290 |
0.1400250958 |
- |
- |
PREDICTED: non-specific phospholipase C1 [Jatropha curcas] |
| 13 |
Hb_000111_390 |
0.1400541525 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001776_200 |
0.1498843941 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 15 |
Hb_002095_050 |
0.1506478269 |
- |
- |
PREDICTED: uncharacterized protein LOC104447479 [Eucalyptus grandis] |
| 16 |
Hb_132840_020 |
0.1532542892 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 17 |
Hb_000086_640 |
0.1575666423 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 18 |
Hb_000748_110 |
0.1583025446 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 19 |
Hb_011671_110 |
0.1621513271 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 20 |
Hb_005724_050 |
0.1650382006 |
- |
- |
- |