| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
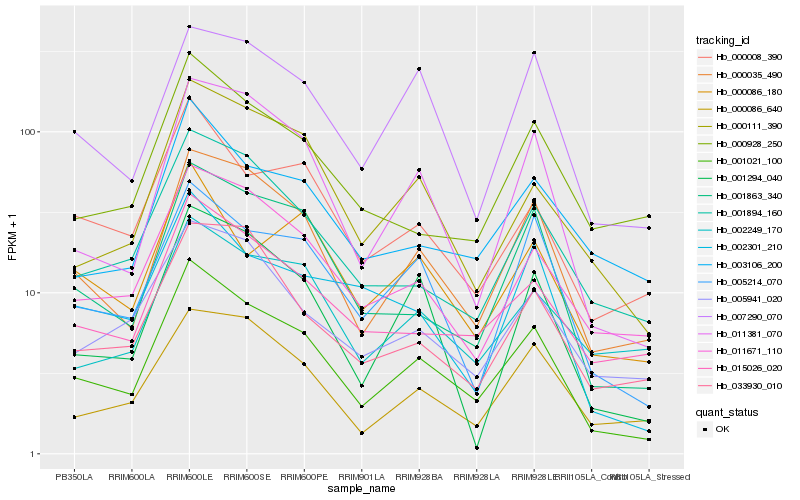
Hb_001021_100 |
0.0 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 2 |
Hb_000035_490 |
0.0930478221 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 3 |
Hb_001863_340 |
0.1151730493 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 4 |
Hb_011381_070 |
0.1161840985 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 5 |
Hb_011671_110 |
0.1216505908 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 6 |
Hb_005214_070 |
0.122055467 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |
| 7 |
Hb_015026_020 |
0.1253114125 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 8 |
Hb_001894_160 |
0.1284963851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000111_390 |
0.129739908 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000008_390 |
0.1298821114 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 11 |
Hb_000086_640 |
0.1372880289 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 12 |
Hb_005941_020 |
0.1382798536 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 13 |
Hb_001294_040 |
0.1395755525 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 14 |
Hb_003106_200 |
0.1406655175 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007290_070 |
0.1473557041 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 16 |
Hb_002249_170 |
0.1487804042 |
- |
- |
copine, putative [Ricinus communis] |
| 17 |
Hb_033930_010 |
0.1491917096 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 18 |
Hb_002301_210 |
0.1495549702 |
- |
- |
PREDICTED: receptor-like protein kinase At5g59670 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000928_250 |
0.1499193782 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_000086_180 |
0.1519282244 |
- |
- |
hypothetical protein JCGZ_17674 [Jatropha curcas] |