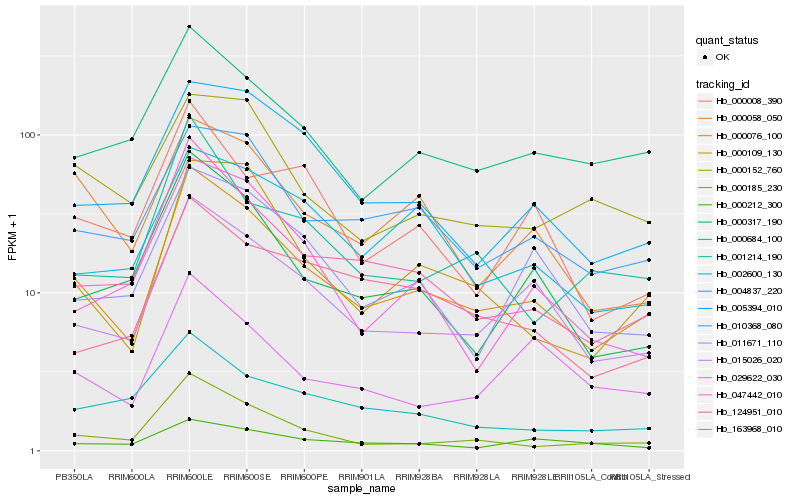
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000076_100 |
0.0 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1 [Jatropha curcas] |
| 2 |
Hb_015026_020 |
0.1060321468 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 3 |
Hb_124951_010 |
0.111528184 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 4 |
Hb_010368_080 |
0.1295633149 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |
| 5 |
Hb_000109_130 |
0.1301635806 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 [Jatropha curcas] |
| 6 |
Hb_000684_100 |
0.1318450451 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 7 |
Hb_000317_190 |
0.1352400829 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 8 |
Hb_005394_010 |
0.140878343 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 9 |
Hb_000185_230 |
0.1420676297 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 10 |
Hb_011671_110 |
0.1447243287 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 11 |
Hb_000058_050 |
0.1448948681 |
- |
- |
PREDICTED: uncharacterized protein LOC105640877 [Jatropha curcas] |
| 12 |
Hb_004837_220 |
0.1530728292 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 13 |
Hb_002600_130 |
0.1547989902 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |
| 14 |
Hb_000152_760 |
0.1552963542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_047442_010 |
0.1566612115 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 16 |
Hb_001214_190 |
0.1566847509 |
- |
- |
hypothetical protein JCGZ_25001 [Jatropha curcas] |
| 17 |
Hb_163968_010 |
0.1574972562 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 18 |
Hb_029622_030 |
0.1578866436 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000008_390 |
0.1621925267 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 20 |
Hb_000212_300 |
0.1628843017 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |