| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
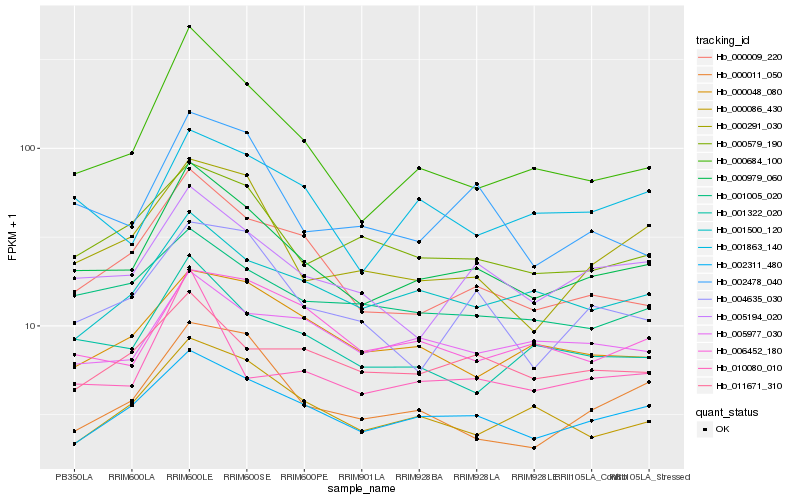
Hb_000979_060 |
0.0 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 2 |
Hb_000684_100 |
0.0934969921 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 3 |
Hb_005194_020 |
0.0939742518 |
- |
- |
PREDICTED: UPF0553 protein-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_002311_480 |
0.094744812 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000009_220 |
0.0981574424 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_001322_020 |
0.1037503991 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 7 |
Hb_000579_190 |
0.1101543503 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 8 |
Hb_000291_030 |
0.1124424267 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 9 |
Hb_002478_040 |
0.1127796472 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2C, putative [Ricinus communis] |
| 10 |
Hb_001500_120 |
0.1140946738 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_000086_430 |
0.1148463844 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 12 |
Hb_001005_020 |
0.1159880491 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 13 |
Hb_001863_140 |
0.1170982258 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 6b-1 [Jatropha curcas] |
| 14 |
Hb_004635_030 |
0.1176174947 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 15 |
Hb_000011_050 |
0.1199670721 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 16 |
Hb_000048_080 |
0.122735917 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006452_180 |
0.1243709606 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 18 |
Hb_011671_310 |
0.1250285258 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 19 |
Hb_010080_010 |
0.1258036467 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 20 |
Hb_005977_030 |
0.1262474954 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |