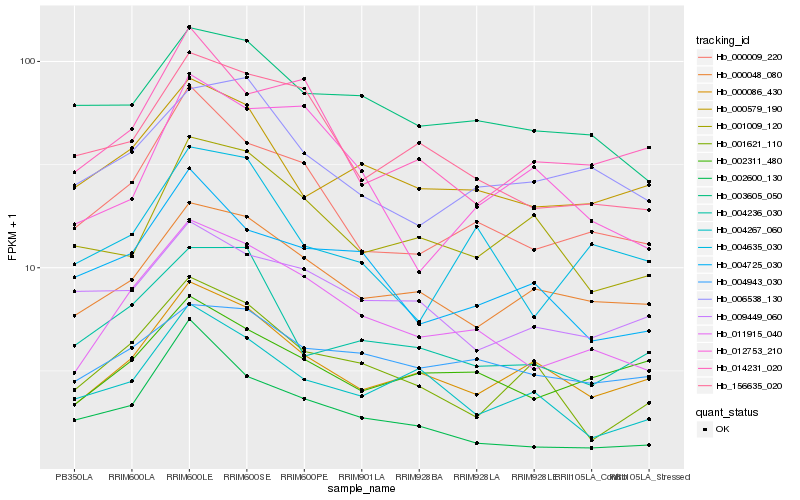
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011915_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 2 |
Hb_000009_220 |
0.1019027272 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_156635_020 |
0.1053916305 |
- |
- |
PREDICTED: L-lactate dehydrogenase B [Jatropha curcas] |
| 4 |
Hb_000048_080 |
0.1236360703 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000086_430 |
0.1281841765 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 6 |
Hb_004943_030 |
0.1308523142 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 7 |
Hb_003605_050 |
0.1316160722 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_004725_030 |
0.1351468227 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |
| 9 |
Hb_006538_130 |
0.1392534005 |
- |
- |
Uncharacterized protein TCM_009867 [Theobroma cacao] |
| 10 |
Hb_002311_480 |
0.1397690729 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000579_190 |
0.1400826932 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 12 |
Hb_002600_130 |
0.1402548094 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |
| 13 |
Hb_014231_020 |
0.1422163786 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 14 |
Hb_004635_030 |
0.1436035398 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 15 |
Hb_004236_030 |
0.1443009788 |
- |
- |
unknown [Lotus japonicus] |
| 16 |
Hb_001621_110 |
0.1457178427 |
- |
- |
PREDICTED: uncharacterized protein LOC105647362 [Jatropha curcas] |
| 17 |
Hb_012753_210 |
0.1465501269 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 18 |
Hb_004267_060 |
0.1469347956 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_009449_060 |
0.1494060792 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 20 |
Hb_001009_120 |
0.1499535348 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |