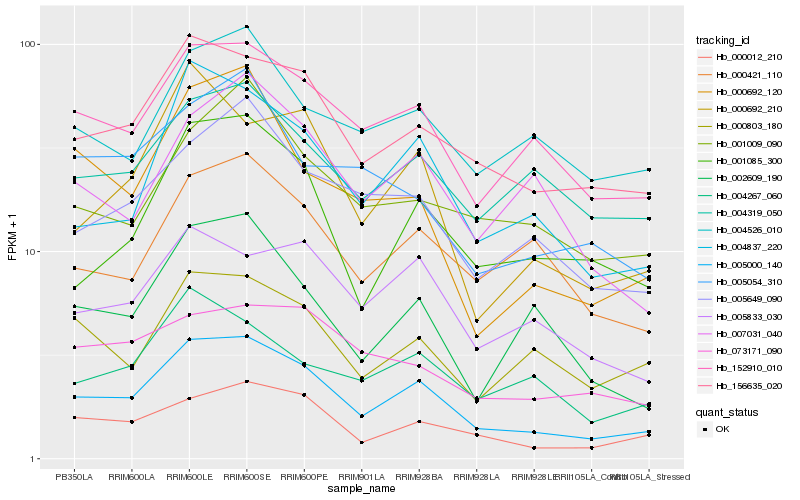
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005000_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637876 isoform X1 [Jatropha curcas] |
| 2 |
Hb_156635_020 |
0.1001395355 |
- |
- |
PREDICTED: L-lactate dehydrogenase B [Jatropha curcas] |
| 3 |
Hb_000012_210 |
0.1073557103 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_152910_010 |
0.1108177808 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 5 |
Hb_004837_220 |
0.1157361325 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 6 |
Hb_005649_090 |
0.1269697057 |
- |
- |
PREDICTED: ninja-family protein AFP3-like [Jatropha curcas] |
| 7 |
Hb_001085_300 |
0.1344597672 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 8 |
Hb_005833_030 |
0.1356102892 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860 [Jatropha curcas] |
| 9 |
Hb_000692_210 |
0.136341967 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2 [Jatropha curcas] |
| 10 |
Hb_002609_190 |
0.1366017872 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 11 |
Hb_000803_180 |
0.1374475929 |
- |
- |
hexokinase [Manihot esculenta] |
| 12 |
Hb_000421_110 |
0.1374718683 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000692_120 |
0.1397581086 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 14 |
Hb_004319_050 |
0.1398037551 |
- |
- |
PREDICTED: senescence-associated carboxylesterase 101-like [Populus euphratica] |
| 15 |
Hb_007031_040 |
0.1402649125 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 16 |
Hb_004267_060 |
0.1432644616 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_001009_090 |
0.1434244512 |
- |
- |
CDK protein [Hevea brasiliensis] |
| 18 |
Hb_073171_090 |
0.1441040667 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 19 |
Hb_005054_310 |
0.1452663251 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 20 |
Hb_004526_010 |
0.1454634767 |
- |
- |
signal recognition particle receptor alpha subunit, putative [Ricinus communis] |