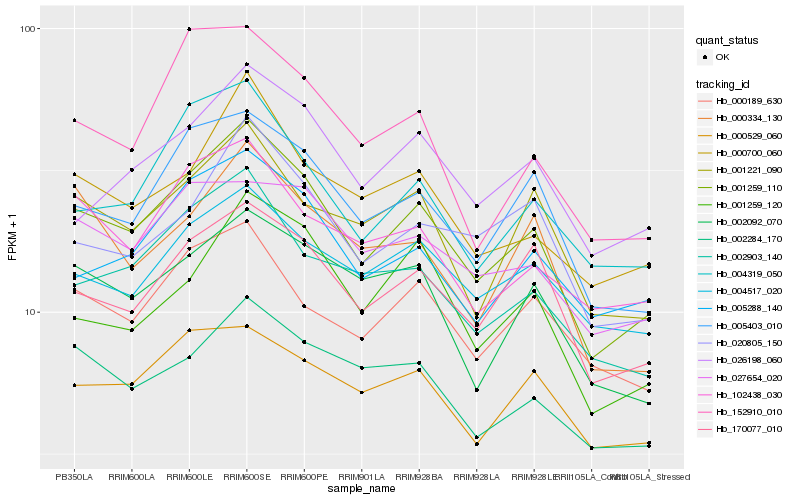
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001259_110 |
0.0 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 3 [Jatropha curcas] |
| 2 |
Hb_005403_010 |
0.0679841669 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 3 |
Hb_170077_010 |
0.0745669906 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002903_140 |
0.0777156405 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 5 |
Hb_000700_060 |
0.0792380157 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 6 |
Hb_001221_090 |
0.0813050356 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005288_140 |
0.0822264002 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 8 |
Hb_004319_050 |
0.0823545744 |
- |
- |
PREDICTED: senescence-associated carboxylesterase 101-like [Populus euphratica] |
| 9 |
Hb_002284_170 |
0.0831912386 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 10 |
Hb_002092_070 |
0.0834106283 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 11 |
Hb_000189_630 |
0.0842231678 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 12 |
Hb_102438_030 |
0.0844792954 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_027654_020 |
0.0858629924 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 14 |
Hb_020805_150 |
0.0864007115 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_004517_020 |
0.0876486833 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 16 |
Hb_001259_120 |
0.0877794698 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 17 |
Hb_026198_060 |
0.0882845765 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |
| 18 |
Hb_152910_010 |
0.0895536969 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 19 |
Hb_000529_060 |
0.0901001506 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000334_130 |
0.0902586006 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |