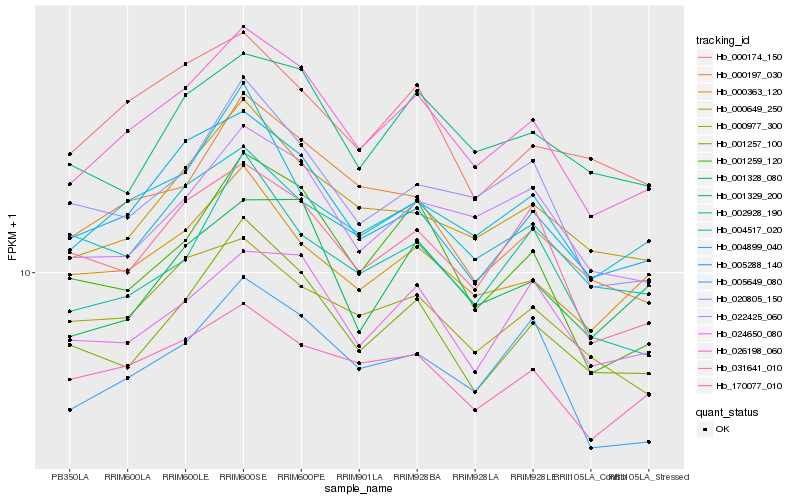
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026198_060 |
0.0 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |
| 2 |
Hb_005288_140 |
0.0691511902 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 3 |
Hb_000649_250 |
0.0708999664 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 4 |
Hb_022425_060 |
0.0731922931 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 5 |
Hb_001257_100 |
0.0746508863 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 6 |
Hb_004517_020 |
0.0753518698 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 7 |
Hb_001259_120 |
0.0756533968 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 8 |
Hb_005649_080 |
0.0760005794 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 9 |
Hb_170077_010 |
0.0772419704 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 10 |
Hb_031641_010 |
0.0794090199 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001329_200 |
0.0807961882 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 12 |
Hb_020805_150 |
0.0820743564 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_002928_190 |
0.0820994778 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 14 |
Hb_004899_040 |
0.0822289301 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 15 |
Hb_024650_080 |
0.0824995637 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 16 |
Hb_000174_150 |
0.0833392531 |
- |
- |
stearoyl-acyl-carrier protein desaturase [Ricinus communis] |
| 17 |
Hb_000977_300 |
0.0839244284 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 18 |
Hb_000363_120 |
0.0845760348 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 19 |
Hb_000197_030 |
0.0850533692 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 20 |
Hb_001328_080 |
0.0861893071 |
- |
- |
PREDICTED: importin subunit alpha-1b [Jatropha curcas] |