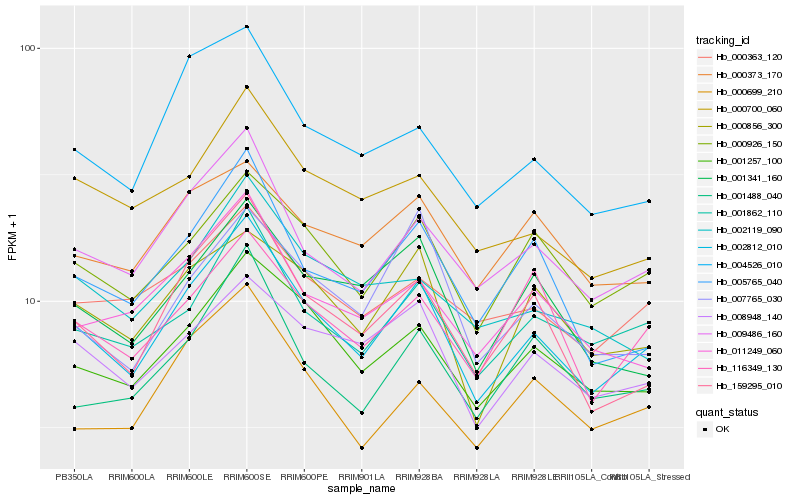
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002812_010 |
0.0 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_009486_160 |
0.0725312151 |
- |
- |
PREDICTED: metacaspase-1 [Jatropha curcas] |
| 3 |
Hb_001257_100 |
0.0746867857 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 4 |
Hb_001862_110 |
0.0790910177 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 5 |
Hb_159295_010 |
0.0791637302 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 6 |
Hb_000926_150 |
0.081944267 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_008948_140 |
0.0887231767 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 8 |
Hb_005765_040 |
0.0895363981 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 9 |
Hb_000856_300 |
0.0931735096 |
- |
- |
PREDICTED: uncharacterized protein LOC105640498 [Jatropha curcas] |
| 10 |
Hb_004526_010 |
0.09321239 |
- |
- |
signal recognition particle receptor alpha subunit, putative [Ricinus communis] |
| 11 |
Hb_011249_060 |
0.0950380627 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 12 |
Hb_000699_210 |
0.0957359511 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 13 |
Hb_000700_060 |
0.0958904281 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 14 |
Hb_001341_160 |
0.0963890057 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001488_040 |
0.0980552742 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 16 |
Hb_000363_120 |
0.1006120774 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 17 |
Hb_116349_130 |
0.1025647569 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000373_170 |
0.1033710549 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 19 |
Hb_007765_030 |
0.1035708803 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 20 |
Hb_002119_090 |
0.1042751435 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |