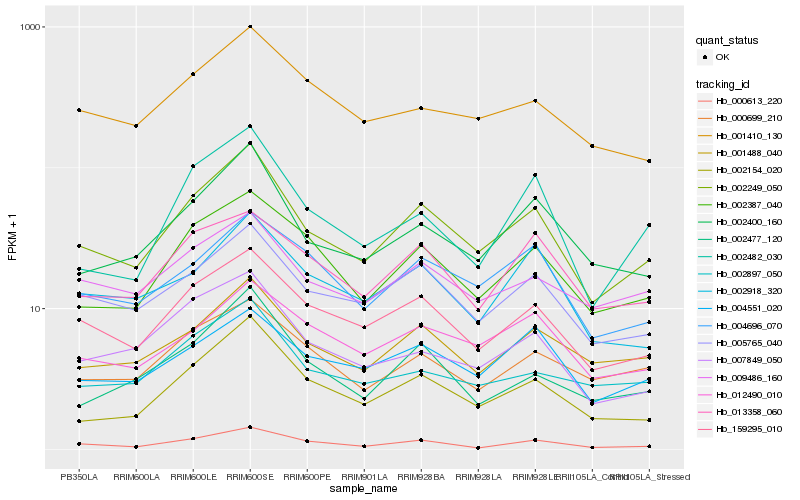
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002249_050 |
0.0 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 2 |
Hb_159295_010 |
0.0812666215 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 3 |
Hb_005765_040 |
0.0878524444 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 4 |
Hb_002400_160 |
0.0915933725 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 5 |
Hb_002154_020 |
0.094841164 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_000613_220 |
0.0974731022 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 7 |
Hb_001488_040 |
0.0996941461 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 8 |
Hb_002387_040 |
0.1002925387 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 9 |
Hb_009486_160 |
0.1112997122 |
- |
- |
PREDICTED: metacaspase-1 [Jatropha curcas] |
| 10 |
Hb_004696_070 |
0.1132635065 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 11 |
Hb_012490_010 |
0.1141243828 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |
| 12 |
Hb_007849_050 |
0.1154290792 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 13 |
Hb_002897_050 |
0.115659531 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 14 |
Hb_000699_210 |
0.1158281159 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 15 |
Hb_002918_320 |
0.1172226812 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004551_020 |
0.1177970455 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 17 |
Hb_002477_120 |
0.1188049911 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 18 |
Hb_002482_030 |
0.1189007857 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |
| 19 |
Hb_001410_130 |
0.1203105152 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 20 |
Hb_013358_060 |
0.120321674 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |