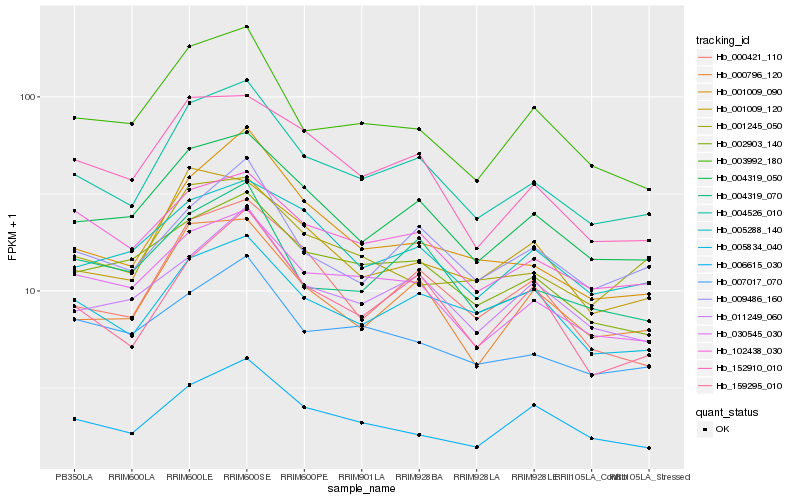
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004526_010 |
0.0 |
- |
- |
signal recognition particle receptor alpha subunit, putative [Ricinus communis] |
| 2 |
Hb_004319_050 |
0.0607698342 |
- |
- |
PREDICTED: senescence-associated carboxylesterase 101-like [Populus euphratica] |
| 3 |
Hb_000796_120 |
0.0723050592 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 4 |
Hb_030545_030 |
0.0727947871 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 5 |
Hb_003992_180 |
0.0803997939 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009486_160 |
0.0821962211 |
- |
- |
PREDICTED: metacaspase-1 [Jatropha curcas] |
| 7 |
Hb_159295_010 |
0.0831181356 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 8 |
Hb_152910_010 |
0.0831740332 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 9 |
Hb_011249_060 |
0.0833674967 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 10 |
Hb_002903_140 |
0.0836759336 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 11 |
Hb_001009_090 |
0.0836896621 |
- |
- |
CDK protein [Hevea brasiliensis] |
| 12 |
Hb_004319_070 |
0.083707986 |
- |
- |
hypothetical protein RCOM_0852460 [Ricinus communis] |
| 13 |
Hb_005834_040 |
0.0854346193 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 14 |
Hb_000421_110 |
0.0854549953 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_102438_030 |
0.0866986243 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_007017_070 |
0.0880034123 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001009_120 |
0.0891289665 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 18 |
Hb_005288_140 |
0.0909902854 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 19 |
Hb_006615_030 |
0.0916604869 |
- |
- |
prolyl oligopeptidase family protein [Populus trichocarpa] |
| 20 |
Hb_001245_050 |
0.0926385592 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |