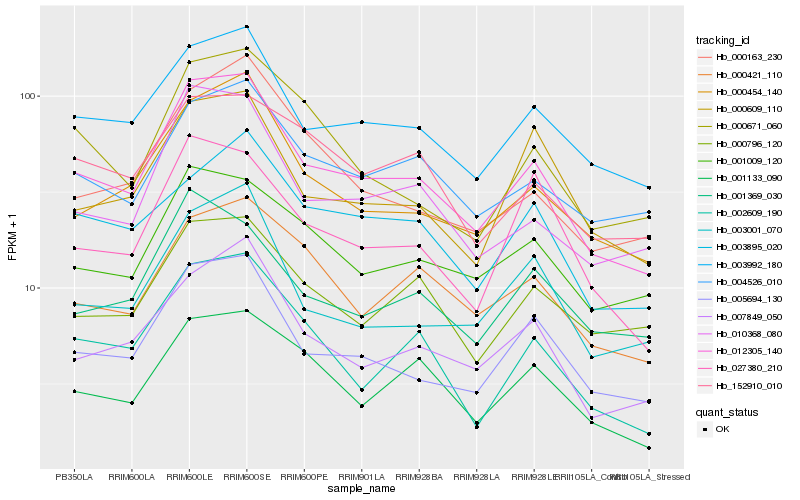
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012305_140 |
0.0 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_005694_130 |
0.0749348492 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 3 |
Hb_003992_180 |
0.0770101111 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000454_140 |
0.0875557533 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2a-like [Jatropha curcas] |
| 5 |
Hb_010368_080 |
0.0895348955 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |
| 6 |
Hb_007849_050 |
0.0916878017 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 7 |
Hb_002609_190 |
0.0992848688 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 8 |
Hb_004526_010 |
0.099572021 |
- |
- |
signal recognition particle receptor alpha subunit, putative [Ricinus communis] |
| 9 |
Hb_001009_120 |
0.1007952736 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000421_110 |
0.1027355352 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001133_090 |
0.1029364591 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 12 |
Hb_003001_070 |
0.1062096127 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0002s24750g [Populus trichocarpa] |
| 13 |
Hb_003895_020 |
0.1081072057 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000609_110 |
0.108821706 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 15 |
Hb_027380_210 |
0.110099902 |
- |
- |
PREDICTED: uncharacterized protein LOC105634030 [Jatropha curcas] |
| 16 |
Hb_000671_060 |
0.1102017886 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_000796_120 |
0.1103019193 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 18 |
Hb_000163_230 |
0.1107338812 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 19 |
Hb_001369_030 |
0.1110385689 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 20 |
Hb_152910_010 |
0.1113606812 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |