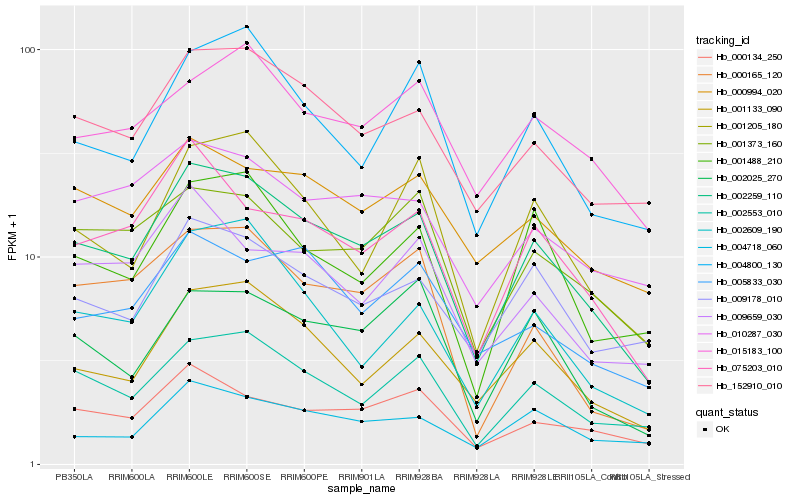
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002259_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 2 |
Hb_002553_010 |
0.0897775696 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 2-like [Populus euphratica] |
| 3 |
Hb_152910_010 |
0.0923752514 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 4 |
Hb_001133_090 |
0.0979610358 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 5 |
Hb_075203_010 |
0.0986979155 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000134_250 |
0.1047229313 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_001373_160 |
0.1082625027 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 8 |
Hb_001205_180 |
0.110248712 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 9 |
Hb_000994_020 |
0.1105546302 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_004800_130 |
0.1116853731 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 11 |
Hb_009178_010 |
0.1121006634 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 12 |
Hb_001488_210 |
0.1133221152 |
- |
- |
PREDICTED: protein RIK [Jatropha curcas] |
| 13 |
Hb_010287_030 |
0.1151848713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002609_190 |
0.1171359929 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 15 |
Hb_002025_270 |
0.1180399443 |
- |
- |
PREDICTED: uncharacterized protein At4g15970 isoform X1 [Jatropha curcas] |
| 16 |
Hb_009659_030 |
0.1192160593 |
- |
- |
PREDICTED: beta-galactosidase 17 [Jatropha curcas] |
| 17 |
Hb_004718_060 |
0.1192192675 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000165_120 |
0.119387573 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 19 |
Hb_005833_030 |
0.1194339027 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860 [Jatropha curcas] |
| 20 |
Hb_015183_100 |
0.1202419292 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |