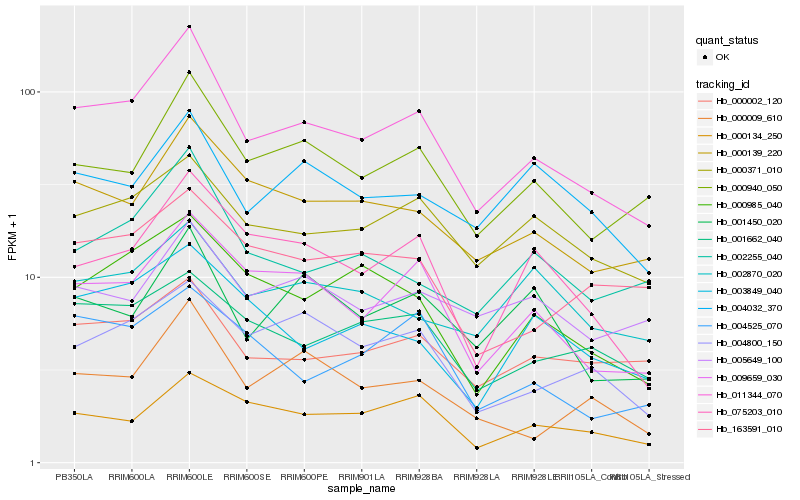
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011344_070 |
0.0 |
- |
- |
PREDICTED: non-specific lipid-transfer protein [Jatropha curcas] |
| 2 |
Hb_009659_030 |
0.1008712633 |
- |
- |
PREDICTED: beta-galactosidase 17 [Jatropha curcas] |
| 3 |
Hb_000139_220 |
0.1064699785 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000940_050 |
0.1091394265 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 5 |
Hb_075203_010 |
0.1216576152 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004800_150 |
0.1229472706 |
transcription factor |
TF Family: SNF2 |
PREDICTED: ATP-dependent DNA helicase DDM1 [Jatropha curcas] |
| 7 |
Hb_000985_040 |
0.1234733175 |
transcription factor |
TF Family: Orphans |
B-box type zinc finger protein with CCT domain isoform 1 [Theobroma cacao] |
| 8 |
Hb_000002_120 |
0.1237847548 |
- |
- |
PREDICTED: uncharacterized protein LOC105782064 [Gossypium raimondii] |
| 9 |
Hb_000371_010 |
0.1241382515 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 10 |
Hb_002255_040 |
0.1266640135 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 11 |
Hb_001662_040 |
0.1274547935 |
- |
- |
DNA helicase, putative [Ricinus communis] |
| 12 |
Hb_000134_250 |
0.1322593305 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_000009_610 |
0.1328951741 |
- |
- |
PREDICTED: uncharacterized protein LOC105639638 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001450_020 |
0.134743276 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_163591_010 |
0.1384755462 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Populus euphratica] |
| 16 |
Hb_003849_040 |
0.1385584378 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 17 |
Hb_005649_100 |
0.1401933176 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 18 |
Hb_004525_070 |
0.141840498 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 19 |
Hb_004032_370 |
0.1425318122 |
- |
- |
pyrophosphate-dependent phosphofructokinase [Hevea brasiliensis] |
| 20 |
Hb_002870_020 |
0.1459290036 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |