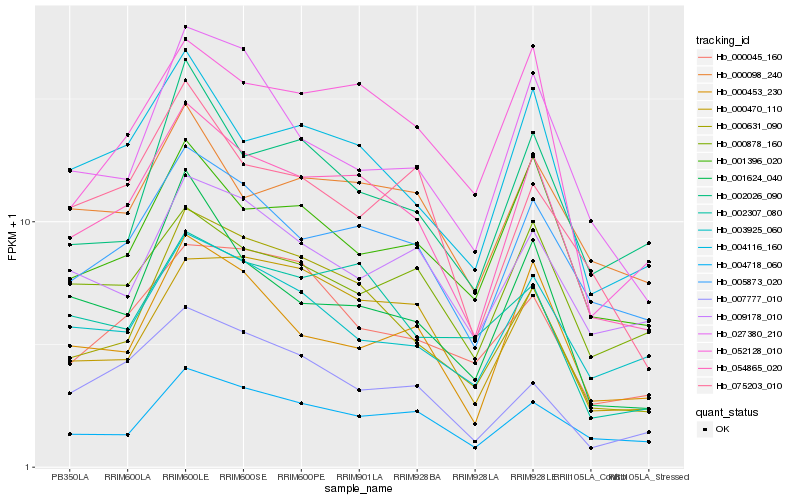
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054865_020 |
0.0 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 2 |
Hb_005873_020 |
0.0733635412 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 3 |
Hb_004116_160 |
0.0843971085 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000631_090 |
0.1051097929 |
- |
- |
PREDICTED: uncharacterized protein LOC105640619 [Jatropha curcas] |
| 5 |
Hb_052128_010 |
0.1051432313 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Nicotiana sylvestris] |
| 6 |
Hb_004718_060 |
0.1076718 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000098_240 |
0.1082366695 |
- |
- |
PREDICTED: UDP-glucose:glycoprotein glucosyltransferase [Jatropha curcas] |
| 8 |
Hb_000453_230 |
0.1126672664 |
- |
- |
PREDICTED: NO-associated protein 1, chloroplastic/mitochondrial [Jatropha curcas] |
| 9 |
Hb_001396_020 |
0.1139438533 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 10 |
Hb_000470_110 |
0.1146753881 |
- |
- |
PREDICTED: probable LIM domain-containing serine/threonine-protein kinase DDB_G0287001 [Jatropha curcas] |
| 11 |
Hb_007777_010 |
0.117059237 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Tarenaya hassleriana] |
| 12 |
Hb_000878_160 |
0.1174368163 |
- |
- |
PREDICTED: D-lactate dehydrogenase [cytochrome], mitochondrial [Jatropha curcas] |
| 13 |
Hb_002307_080 |
0.117927864 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003925_060 |
0.1182634088 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001624_040 |
0.1207334849 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X1 [Jatropha curcas] |
| 16 |
Hb_009178_010 |
0.1208918443 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 17 |
Hb_027380_210 |
0.122868623 |
- |
- |
PREDICTED: uncharacterized protein LOC105634030 [Jatropha curcas] |
| 18 |
Hb_075203_010 |
0.1239796735 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002026_090 |
0.1240072191 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |
| 20 |
Hb_000045_160 |
0.1266553113 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |