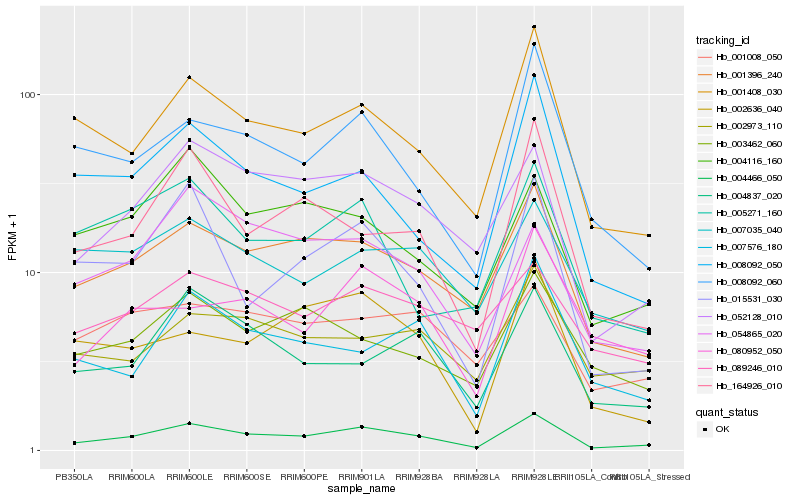
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004466_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 2 |
Hb_001396_240 |
0.1082018112 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_052128_010 |
0.1167643227 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Nicotiana sylvestris] |
| 4 |
Hb_001408_030 |
0.1264452784 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |
| 5 |
Hb_008092_060 |
0.1445750646 |
- |
- |
- |
| 6 |
Hb_015531_030 |
0.1453742497 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_164926_010 |
0.1478430873 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 8 |
Hb_004837_020 |
0.1492275402 |
- |
- |
PREDICTED: uncharacterized protein LOC105648271 [Jatropha curcas] |
| 9 |
Hb_002973_110 |
0.1520799182 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_008092_050 |
0.1526223341 |
- |
- |
PREDICTED: uncharacterized protein LOC105142434 [Populus euphratica] |
| 11 |
Hb_054865_020 |
0.1534415008 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 12 |
Hb_080952_050 |
0.153826946 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Nelumbo nucifera] |
| 13 |
Hb_004116_160 |
0.1546267661 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_007576_180 |
0.1546424149 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 15 |
Hb_002636_040 |
0.1558789576 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001008_050 |
0.1561753189 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 17 |
Hb_003462_060 |
0.1562187543 |
- |
- |
PREDICTED: ABC transporter G family member 24-like [Jatropha curcas] |
| 18 |
Hb_089246_010 |
0.158424134 |
- |
- |
hypothetical protein JCGZ_24395 [Jatropha curcas] |
| 19 |
Hb_005271_160 |
0.1589038108 |
- |
- |
PREDICTED: uncharacterized protein LOC105645844 [Jatropha curcas] |
| 20 |
Hb_007035_040 |
0.159041484 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |