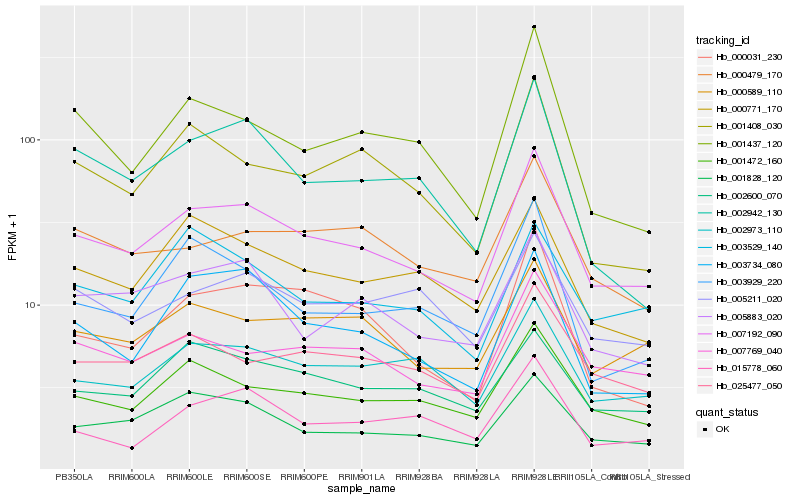
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007192_090 |
0.0 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 2 |
Hb_001472_160 |
0.084321281 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 3 |
Hb_001828_120 |
0.0896471936 |
- |
- |
PREDICTED: uncharacterized protein LOC105140167 [Populus euphratica] |
| 4 |
Hb_007769_040 |
0.0929983453 |
- |
- |
PREDICTED: probable threonine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 5 |
Hb_002973_110 |
0.095495295 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_025477_050 |
0.0976914372 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_000589_110 |
0.0983223036 |
- |
- |
PREDICTED: uncharacterized protein LOC105647614 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005883_020 |
0.1024161343 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74850, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000479_170 |
0.1063064192 |
- |
- |
chaperone clpb, putative [Ricinus communis] |
| 10 |
Hb_002600_070 |
0.1071642626 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 11 |
Hb_000771_170 |
0.1073874807 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 12 |
Hb_005211_020 |
0.1092796408 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_003734_080 |
0.1115579775 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 14 |
Hb_003929_220 |
0.1140959371 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_001408_030 |
0.1149619979 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001437_120 |
0.1150832368 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Jatropha curcas] |
| 17 |
Hb_002942_130 |
0.115893059 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000031_230 |
0.1178272308 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003529_140 |
0.1193829417 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_015778_060 |
0.1194686351 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |