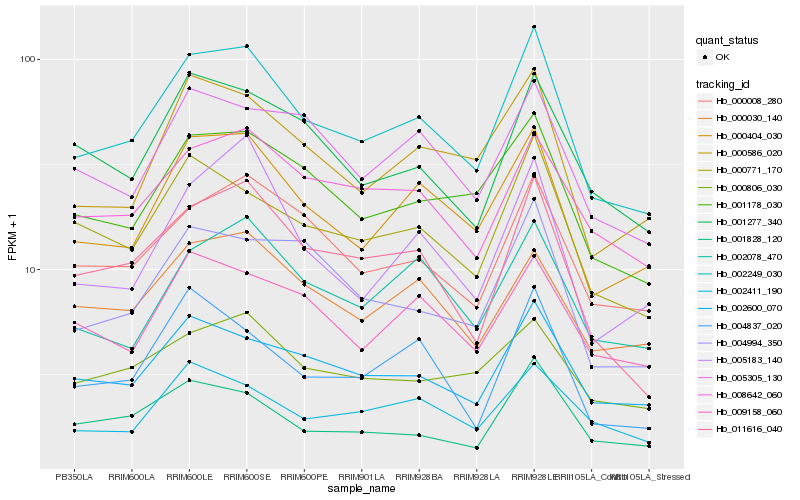
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002249_030 |
0.0 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000404_030 |
0.0692966805 |
- |
- |
PREDICTED: protein NAP1 [Jatropha curcas] |
| 3 |
Hb_011616_040 |
0.0834437112 |
- |
- |
glucan water dikinase 3, partial [Manihot esculenta] |
| 4 |
Hb_001178_030 |
0.0837386178 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 2-2, chloroplastic/amyloplastic [Jatropha curcas] |
| 5 |
Hb_000008_280 |
0.0850507653 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 6 |
Hb_002600_070 |
0.0875717596 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 7 |
Hb_000771_170 |
0.0876185493 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 8 |
Hb_000586_020 |
0.088337101 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein F383_02497 [Gossypium arboreum] |
| 9 |
Hb_008642_060 |
0.0955337317 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 10 |
Hb_002078_470 |
0.0966949349 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004994_350 |
0.0983305836 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001828_120 |
0.0987469729 |
- |
- |
PREDICTED: uncharacterized protein LOC105140167 [Populus euphratica] |
| 13 |
Hb_001277_340 |
0.1001125892 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 14 |
Hb_005305_130 |
0.1009395599 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 15 |
Hb_000030_140 |
0.1020768166 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000806_030 |
0.104818805 |
transcription factor |
TF Family: C2H2 |
PREDICTED: probable lysine-specific demethylase ELF6 [Jatropha curcas] |
| 17 |
Hb_009158_060 |
0.1061301254 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 18 |
Hb_005183_140 |
0.1073232431 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 19 |
Hb_002411_190 |
0.1081211836 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 20 |
Hb_004837_020 |
0.1086766908 |
- |
- |
PREDICTED: uncharacterized protein LOC105648271 [Jatropha curcas] |