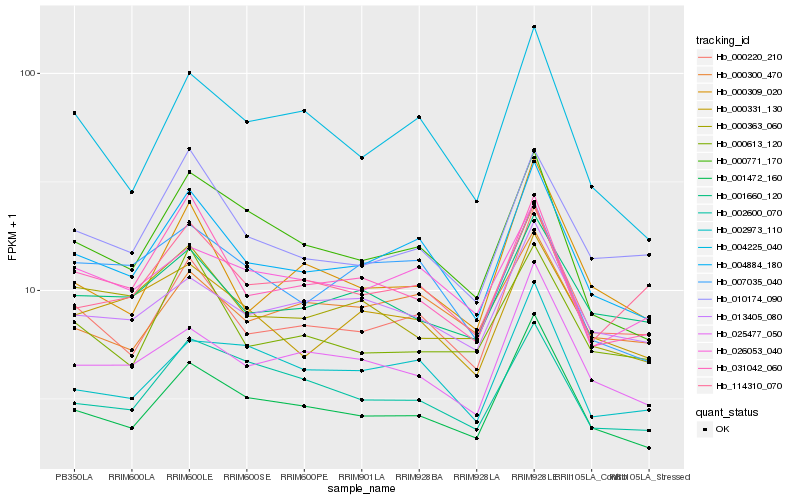
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004884_180 |
0.0 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001472_160 |
0.0768025251 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 3 |
Hb_000220_210 |
0.0795316455 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 4 |
Hb_031042_060 |
0.0834026299 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 5 |
Hb_000771_170 |
0.0839434434 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_010174_090 |
0.086253508 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004225_040 |
0.0864625906 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Populus euphratica] |
| 8 |
Hb_007035_040 |
0.0871984556 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000331_130 |
0.0882550827 |
transcription factor |
TF Family: Trihelix |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000300_470 |
0.0925260281 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 11 |
Hb_026053_040 |
0.0931655169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_025477_050 |
0.0932893783 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_000363_060 |
0.0933166032 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_013405_080 |
0.093978782 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 15 |
Hb_001660_120 |
0.0947209973 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002973_110 |
0.0948895485 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_000309_020 |
0.0972903512 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 18 |
Hb_002600_070 |
0.0983468615 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 19 |
Hb_114310_070 |
0.098622934 |
- |
- |
GTP-dependent nucleic acid-binding protein engD, putative [Ricinus communis] |
| 20 |
Hb_000613_120 |
0.0999553741 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |