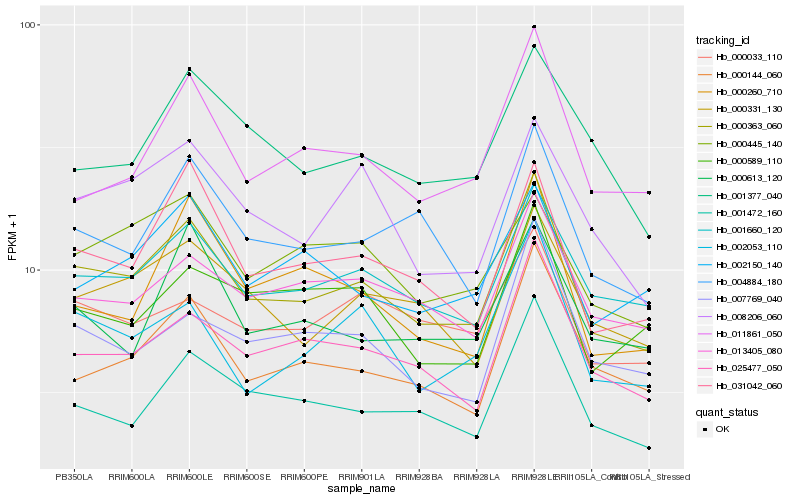
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_060 |
0.0 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001660_120 |
0.074607696 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_025477_050 |
0.0826209754 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_031042_060 |
0.08366257 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 5 |
Hb_001472_160 |
0.088294374 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 6 |
Hb_000331_130 |
0.0896659901 |
transcription factor |
TF Family: Trihelix |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000144_060 |
0.0921027786 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_013405_080 |
0.0924894486 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 9 |
Hb_004884_180 |
0.0933166032 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002053_110 |
0.0943443543 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_008206_060 |
0.0966094464 |
- |
- |
PREDICTED: preprotein translocase subunit SCY1, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_007769_040 |
0.0980790085 |
- |
- |
PREDICTED: probable threonine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 13 |
Hb_011861_050 |
0.0997029161 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_000033_110 |
0.1035806703 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 15 |
Hb_000260_710 |
0.105518195 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001377_040 |
0.1063739143 |
- |
- |
chitinase, putative [Ricinus communis] |
| 17 |
Hb_000613_120 |
0.1068150125 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000589_110 |
0.1069690273 |
- |
- |
PREDICTED: uncharacterized protein LOC105647614 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002150_140 |
0.107205728 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 20 |
Hb_000445_140 |
0.1073169474 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |