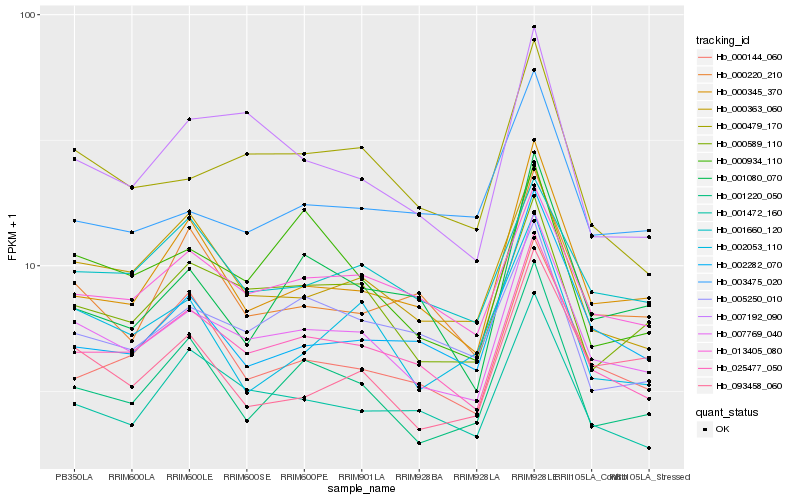
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007769_040 |
0.0 |
- |
- |
PREDICTED: probable threonine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 2 |
Hb_025477_050 |
0.0605672892 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_000589_110 |
0.0828858367 |
- |
- |
PREDICTED: uncharacterized protein LOC105647614 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000345_370 |
0.0869879426 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 5 |
Hb_000479_170 |
0.0874654743 |
- |
- |
chaperone clpb, putative [Ricinus communis] |
| 6 |
Hb_013405_080 |
0.0874863487 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 7 |
Hb_001220_050 |
0.0918030851 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 8 |
Hb_001472_160 |
0.092782068 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 9 |
Hb_007192_090 |
0.0929983453 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 10 |
Hb_000363_060 |
0.0980790085 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000144_060 |
0.0997097267 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_002282_070 |
0.1013424811 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 13 |
Hb_001660_120 |
0.1019540111 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005250_010 |
0.1043624811 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 15 |
Hb_093458_060 |
0.1052743035 |
- |
- |
PREDICTED: uncharacterized protein LOC105633194 [Jatropha curcas] |
| 16 |
Hb_000220_210 |
0.1055447721 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000934_110 |
0.1084693898 |
- |
- |
PREDICTED: protein trichome berefringence-like 7 [Jatropha curcas] |
| 18 |
Hb_003475_020 |
0.1089153041 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 19 |
Hb_001080_070 |
0.1096924278 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 20 |
Hb_002053_110 |
0.1110130541 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |