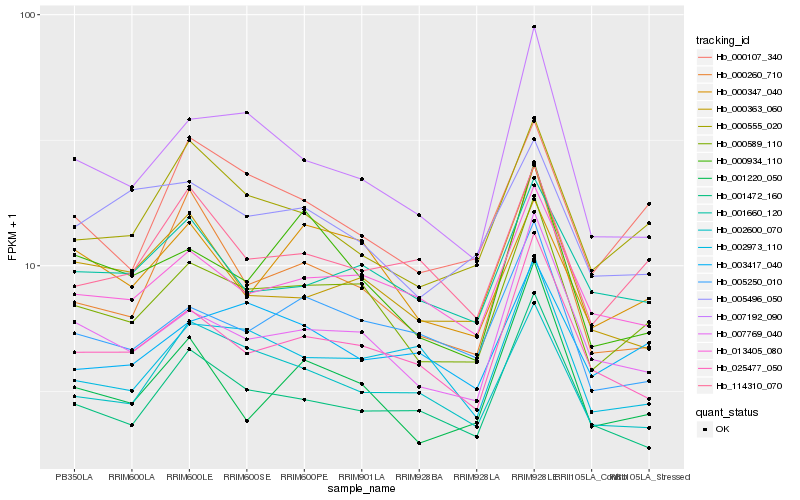
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000589_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647614 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007769_040 |
0.0828858367 |
- |
- |
PREDICTED: probable threonine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 3 |
Hb_013405_080 |
0.0844748855 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 4 |
Hb_005250_010 |
0.0921182215 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 5 |
Hb_025477_050 |
0.0923619677 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_007192_090 |
0.0983223036 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 7 |
Hb_000555_020 |
0.0994765793 |
- |
- |
APO protein 2, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_001660_120 |
0.0997107716 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000347_040 |
0.1031821942 |
- |
- |
hypothetical protein JCGZ_24660 [Jatropha curcas] |
| 10 |
Hb_000934_110 |
0.1031994539 |
- |
- |
PREDICTED: protein trichome berefringence-like 7 [Jatropha curcas] |
| 11 |
Hb_000107_340 |
0.1043606368 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105637652 [Jatropha curcas] |
| 12 |
Hb_000363_060 |
0.1069690273 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_114310_070 |
0.1071709346 |
- |
- |
GTP-dependent nucleic acid-binding protein engD, putative [Ricinus communis] |
| 14 |
Hb_002600_070 |
0.108991061 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 15 |
Hb_003417_040 |
0.1099716899 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 16 |
Hb_001472_160 |
0.1100142308 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 17 |
Hb_001220_050 |
0.1101620526 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 18 |
Hb_005496_050 |
0.1106786041 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 19 |
Hb_002973_110 |
0.110689759 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_000260_710 |
0.1115727128 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |